Bacteriophages consist of a protein coat that surrounds/protects their genomes; the arrangement of the genome inside this coat has been proposed by several researchers. Like any other virus, bacteriophages compose of either Deoxyribonucleic acid (DNA) or Ribonucleic acid (RNA) on their genomes but not both, which may encode for less than ten (MS2 bacteriophage genome encode for four genes ) up to a hundred genes (T4 bacteriophage genome encode for hundreds of genes). Bacteriophage genomes vary greatly in shapes and form; RNA phages tend to have much smaller ones than DNA ones. The bacteriophage genome may be single or double-stranded, linear, or circular.
Bacteriophage genome packaging
The genomes of most tailed phages are contained within the capsid at a ∼500 mg per ml concentration. The DNA inside the capsid is arranged on a closely packed but imperfect hexagonal lattice. Packaging is thought to occur from the outside of the spool inwards. Hydrolysis of Adenosine Triphosphate (ATP) by terminase provides the necessary energy for packaging DNA into the capsid, which may generate a force of up to 100 pN. DNA is highly condensed at this concentration, and much of the water that usually hydrates the DNA and its counterions must be removed to package a complete genome. Water removal during the packaging process occurs by reverse osmosis, the energy derived from the activity of terminase.
Terminase is the critical component in bacteriophage genome packaging machines; this enzyme is usually a heteromultimeric composed of one large (large Ter subunit is an endonuclease/translocase subunit) and one small (small Ter subunit is a recognition subunit) subunit. This enzyme is also found in herpes viruses portraying similar functions.
Only about 10% of the energy stored in the packaged phage DNA is due to bending DNA more tightly than its persistence length. Most of the energy internal to the capsid is due to the dehydrated state of the DNA, which is reflected by internal osmotic pressures that reach tens of atmospheres. The continuum mechanics model posits that the energy stored in the packaged DNA is used to drive the ejection of the phage genome. This model is supported by the experimental suppression of DNA ejection by applying an increased external osmotic pressure Molineux et al. (2013). The hydrodynamic model of phage genome ejection proposes that following the opening of the exit channel, water diffuses through the capsid shell to neutralize the osmotic imbalance, and DNA is pushed out by the hydrostatic pressure gradient across the tail. The data used to support the continuum mechanics model for in vitro ejection is consistent with the hydrodynamic model.
According to the continuum mechanics model, during infection of a bacterial cell, the osmotic pressure of the cytoplasm opposes phage DNA ejection driven by forces internal to the phage capsid; only about half the genome, at most, can enter the cell by this process. The researchers have proposed various ad hoc secondary mechanisms to complete the infection process. The hydrodynamic model has no such problem, as continued water flow up the osmotic gradient (from the growth medium, through the capsid, and into the cytoplasm) provides the necessary force for complete genome transport into the cell.
Bacteriophage genome ejection
For bacteriophages like T4 and P1, which possess a well-structured shape, their genomes reside in the head section of the particle. During infection, bacteriophages expulse their genetic materials via the tail part. A study done in T4 phages by Molineux et al. (2013) suggested a general mechanism of phage genome ejection in vivo involves the width of the exit channel, which determines whether the hydrodynamic flow can facilitate genome ejection. When the channel is wide enough for water and ions to pass through simultaneously as the genome, then the hydrodynamic flow is sufficient; this is likely to be the pathway used by most phage types. However, if the exit channel is only wide enough for double-stranded DNA alone, energy-requiring motors are necessary to translocate the infecting phage genome into the cell.
Thanks for visiting The Phage. if you have liked it and you don’t want to miss out, feel free to subscribe to our newsletter by clicking here
REFERENCES
- Heming JD, Huffman JB, Jones LM, Homa FL. Isolation and characterization of the herpes simplex virus 1 terminase complex. J Virol. 2014 Jan;88(1):225-36. DOI: 10.1128/JVI.02632-13. Epub 2013 Oct 23.
- Rao, V.B., Black, L.W. Structure and assembly of bacteriophage T4 head. Virol J 7, 356 (2010). https://doi.org/10.1186/1743-422X-7-356
- Molineux, I., Panja, D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol 11, 194–204 (2013). https://doi.org/10.1038/nrmicro2988


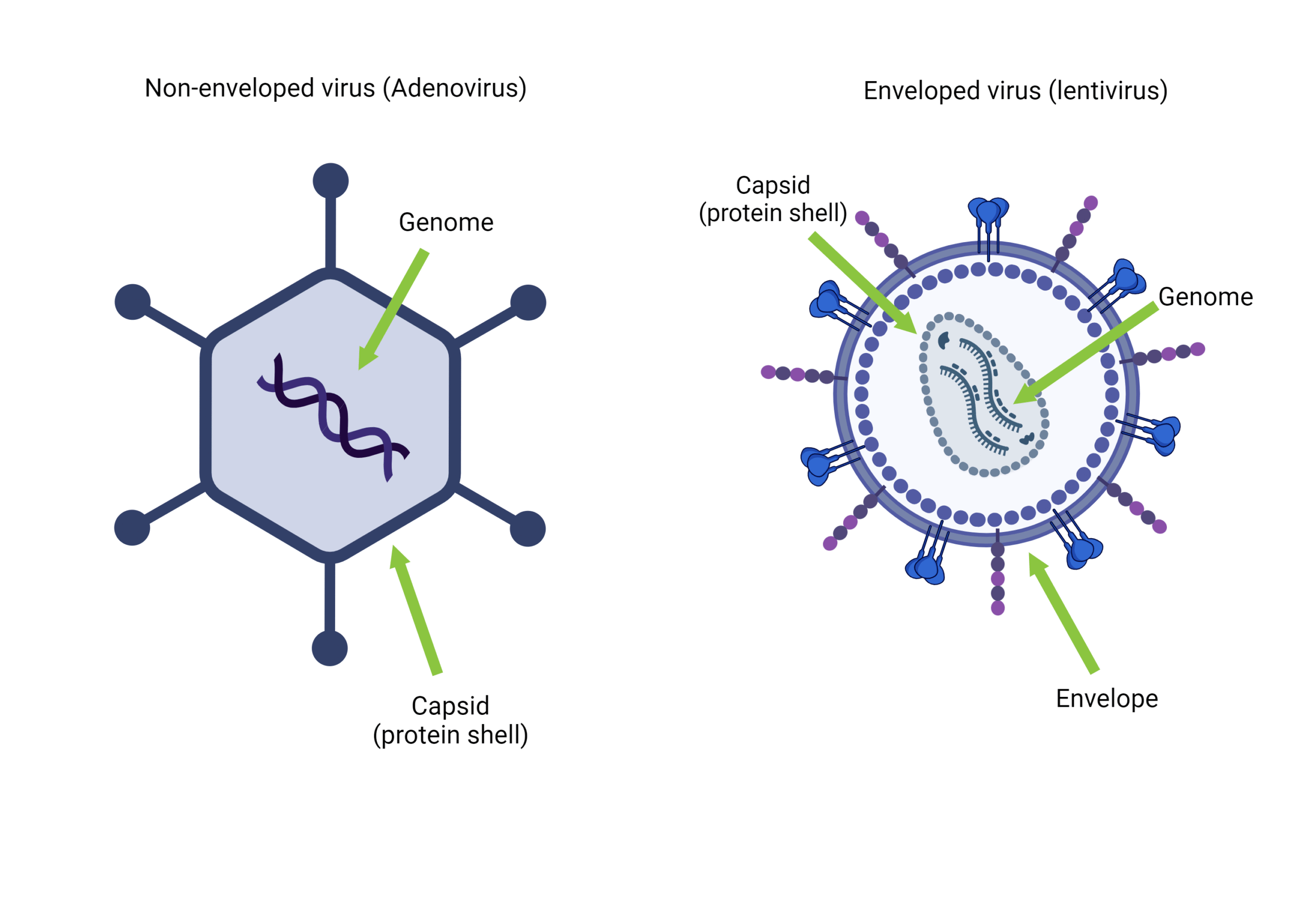
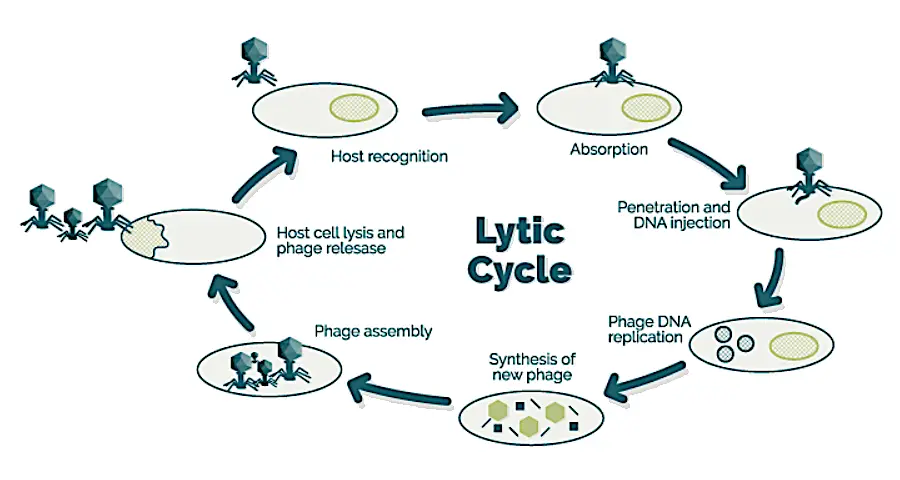
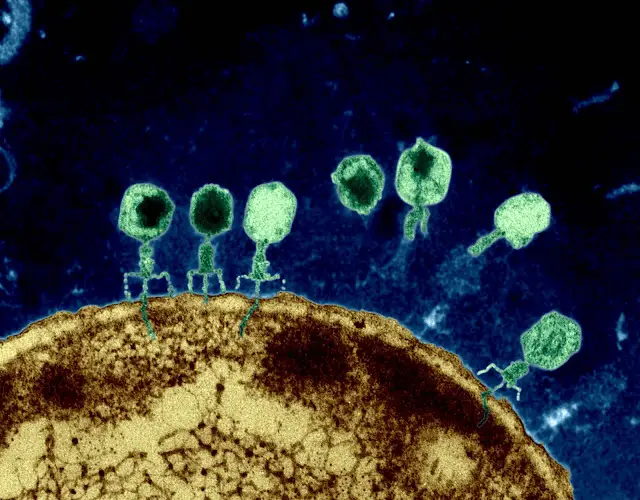
[…] contrast to conventional bacteriophages that inject their entire genomic material into bacterial cells upon infection, T5-like phages, including BF23, employ a distinctive two-stage infection process. […]