 |
| An illustration of ARG transfer by T bacteriophages |
Bacteriophages: viral shuttles carrying bacterial DNA
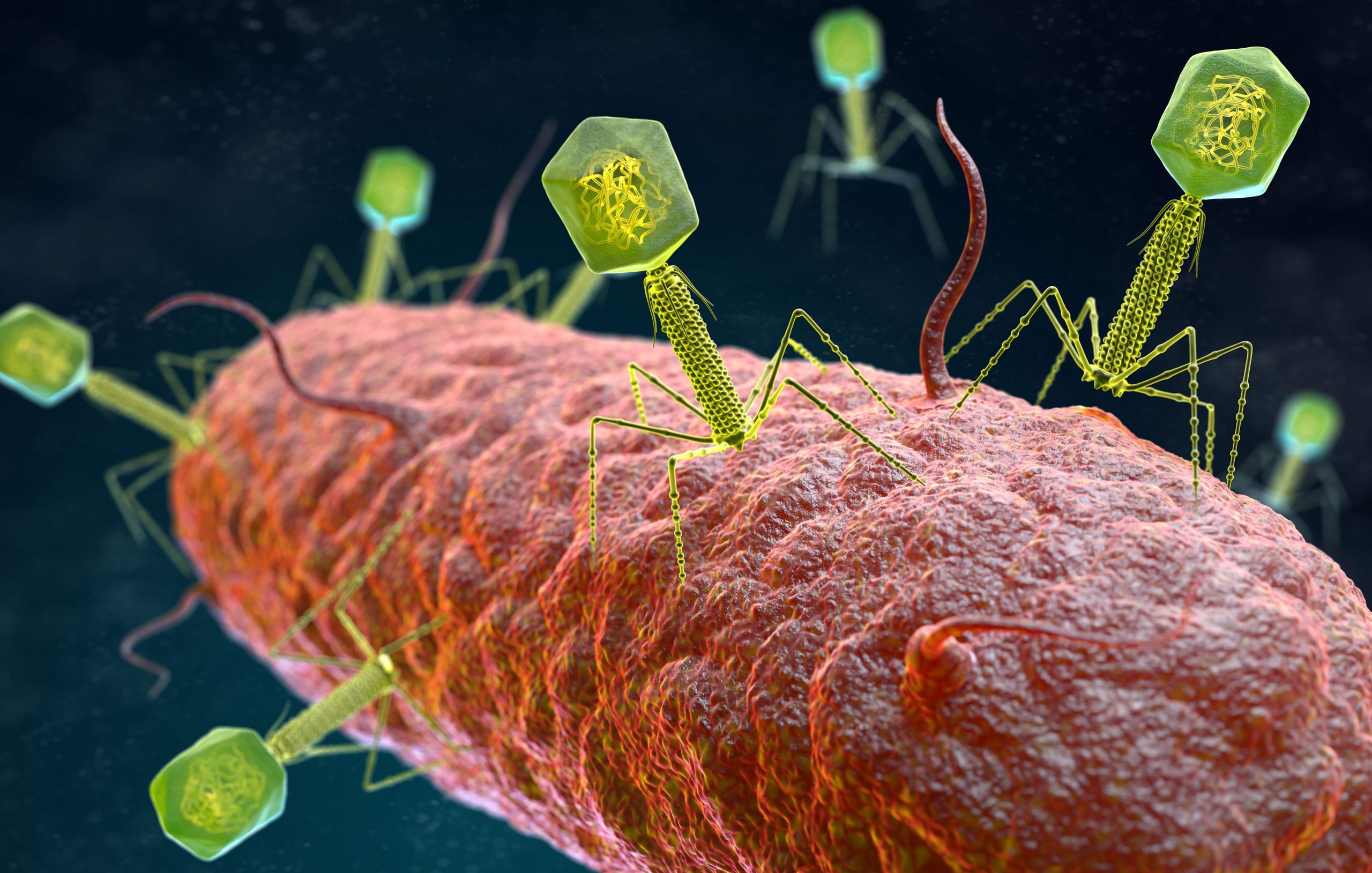
Bacteriophages, like all viruses, are obligate intracellular parasites that rely on the metabolic machinery of the host organism to support their reproduction. A phage’s complete structure typically consists of a nucleic acid core (single or double-stranded RNA or DNA but not both), an outer shell of protein capsid, and, in some instances, a lipid envelope; many of them have additional structures such as the tail, aimed at injecting the nucleic acid through the cell wall) bacteriophages are ultramicroscopic (20–200 nm). As a result, after capsid staining, they must be observed using transmission electron microscopy. In fact, the International Committee on Taxonomy of Viruses (ICTV) taxonomic system requires visualization of the phage particles using electron microscopy, among other characteristics, to determine capsid morphology and phage taxonomy.
Phages are recognized as important players in various aspects of bacterial ecologies, such as bacterial population regulation and, more specifically, gene transfer. Metagenomic studies confirm that the majority of viruses in the viral fraction of most environments are bacteriophages and that a large proportion of viral particles encompass bacterial DNA sequences, including linear chromosome fragments and mobile elements such as insertion elements. By encouraging gene transfer, you can impact the evolution of most bacterial species. Furthermore, phage-like particles that can package random pieces of the host cell’s genome have evolved.
Because bacteriophages are ubiquitous and abundant, gene transfer from phages to bacteria occurs in a diverse range of environments and ecosystems. Since bacteriophage-mediated antibiotic resistance transfer can occur in laboratories, the presence of phages inhabiting ARGs has been reported in a variety of matrices ranging from the human gut to ready-to-eat food. Transduction could play a significant role in the emergence and persistence of antibiotic resistance in these environments, thereby affecting the food chain. In fact, transduction has been identified as a potential contributor to the spread of ARGs, particularly among members of the same species.
If the transferred bacterial DNA recombines into the recipient cell’s genome or guarantees its autonomous replication (i.e., plasmids) transduction has occurred, a capsid containing bacterial DNA can fully bind to the recipient cell and inject foreign DNA.
How do bacteriophages promote ARG dissemination?
Phages bind to specific target receptors on the bacterial cell surface, so each phage typically targets a very narrow range of strains of the same bacterial species. Following adsorption, phages inject their genomes into the bacterial cytoplasm and replicate using one of two primary life cycles: the lytic cycle or the lysogenic (or temperate) cycle.
 |
| bacteriophage cycle |
During the lytic cycle, the phage takes advantage of the host cellular machinery, replicating and manufacturing new phage particles that are released after programmed cell lysis of an overburdened cell. In some cases, pseudo-lysogeny occurs, in which the phage genome remains in nonreplicating cells until they resume replication and the bacteriophage continues its lytic cycle.
The phage genome (known as a prophage) integrates into the bacterial genome during the lysogenic cycle. It replicates as part of the host chromosome or as an independent replicon in the absence of particle formation or cell lysis. The prophage is generated and the lytic cycle is activated under specific conditions that induce the bacterial SOS response (antibiotic treatment, oxidative stress, irradiation, or DNA damage). Host DNA from genes in the prophage integration locus is occasionally incorporated into the phage genome, whereas those that persist as independent replicons contain random fragments.
During the lytic cycle, released phages can package and transfer bacterial DNA at random using generalized transduction. Indeed, some mobile genetic elements have devised sophisticated strategies to exploit the phage DNA packaging machinery for their own transfer. Because phage infection kills bacterial cells during the lytic cycle and small amounts of bacterial DNA are occasionally captured in viral transducing particles, bacterial populations containing prophages can be considered gene transfer drivers, particularly for genes encoding antibiotic resistance.
This context also applies to ARGs; for example, Haaber et al. (2016) demonstrated that the release of phages from a population of S. aureus cells allows the entire prophage-containing population to acquire ARGs from competing, phage susceptible strains present in the same environment. This fact would explain the rapid exchange of ARGs observed in S. aureus and other pathogenic bacteria, despite the fact that bacteriophages are generally thought to infect only
References
- Dennis H Bamford, Jonathan M Grimes, David I Stuart, What does structure tell us about virus evolution?, Current Opinion in Structural Biology, Volume 15, Issue 6, 2005.
- Lerminiaux NA, Cameron ADS. Horizontal transfer of antibiotic resistance genes in clinical environments a J Microbiol 019 Jan;65(1):34-44 oi: 10.1139/cjm-2018-0275. Epub 2018 Sep 24 MID: 30248271.
- Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbé J, Penadés JR. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol 006 Apr;188(7):2726-9 oi: 10.1128/JB.188.7.2726-2729.2006 MID: 16547063; PMCID: PMC1428414.
- Łoś M, Węgrzyn G. Pseudolysogeny. Adv Virus Res. 2012;82:339-49. doi: 10.1016/B978-0-12-394621-8.00019-4. PMID: 22420857.
- Bacteriophages as antibiotic resistance genes carriers in agro‐food systems
S,. JebriF. RahmaniF. Hamid
First published: Sep 11, 2020

These entities have both advantages and disadvantages. we have top be keen on using them
[…] Bacteriophage transduction is the process by which a bacteriophage shuttles or transfers bacterial genes from one bacterial cell to another. The cell whose genome segment is being shuttled is known as the donor cell, and the cell receiving is known as the recipient cell. Transducing phage is the bacteriophage that transfers the bacterial segment. The transduction can be generalized or specialized. Among the genes that bacteria can acquire from phages via transduction are virulence and antibiotic resistance genes. […]
[…] genome. These new genes can bestow advantageous traits upon the recipient cell, such as antibiotic resistance or increased […]