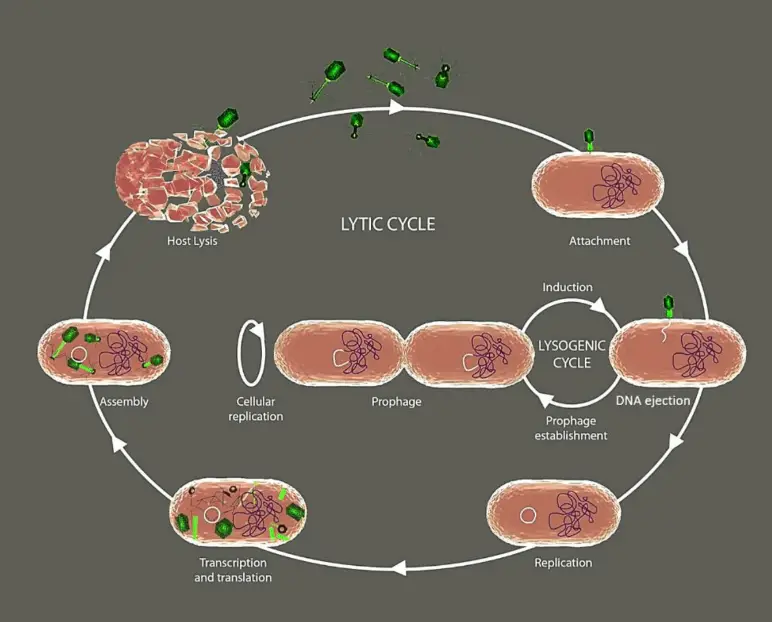
The primary distinction between prophage and bacteriophage is that prophage is a viral genome that has been integrated into a bacterial genome, whereas bacteriophage is the actual virus that infects the bacteria. Prophages are also bacteriophages that go through the lysogenic cycle by integrating their genome into the bacterial genome. Bacteriophages can transform into prophages after integrating their nucleic acids into the bacterial genome. Once integrated, the segment of the genome is known as a prophage. This integration into the host genome is accomplished by an enzyme called integrase, which is encoded by lysogenic bacteriophages. Prophages can undergo excision from the host genome in order to enter into the lytic cycle; research indicates that this process is triggered by external factors.
The enzyme: Integrase
Phage integrases are essential for the incorporation of bacteriophage nucleic acids into the bacterial genome, resulting in the formation of a prophage. These enzymes catalyse unidirectional site-specific recombination between the phage attachment site (attP) and the bacterial attachment site (attB). Based on their mode of catalysis, integrases are divided into two major families: tyrosine recombinases and serine recombinases.
Tyrosine family integrases
Tyrosine family integrases, such as lambda integrase, mediate strand cleavage with a catalytic tyrosine, prefer longer attP sequences, and require the presence of other proteins produced by the phage or the host bacteria.
Structure
The 356 amino acid lambda integrase is made up of two domains: one at the N-terminus that binds the arm-type sites of attP and contains the catalytic site, and one at the C-terminus that binds the lower affinity core-type sites and also contains the catalytic site. The CTD is further subdivided into the core-type binding domain (residues 65-169) and the catalytic core domain (170-356), of which this entry is a part. The catalytic core is folded in an alpha3-beta3-alpha4 pattern, with one side of the beta-sheet exposed.
The tyrosine recombinase family includes the recombinases Cre from phage P1, XerD from E. coli, and Flp from yeast. They all have a two-domain motif similar to lambda integrase and a conserved binding mechanism. Their catalytic core domains have the same structural fold as Lambda integrase.
Serine family integrases
Serine integrases are enzymes that catalyse phage genome integration and excision into and out of bacterial host chromosomes. During viral integration, an integrase (Int) dimer binds to specific ‘attachment site’ sequences in the phage (attP) and host (attB) DNA. The complexes Int-attP and Int-attB form a tetrameric intermediate that cleaves all four substrate DNA strands and covalently couples the Int subunits to their corresponding DNA half-sites. Strand exchange happens when two Int-DNA subunits rotate 180 degrees relative to the other two subunits, and the re-aligned DNA half-sites are ligated to form new attachment sites (attL and attR).
In the absence of a phage-encoded recombination directionality factor (RDF), the integration reaction is effectively unidirectional; the Int-bound attL and attR locations do not efficiently recombine to form attP and attB. Serine integrases are appealing agents in gene therapy and genomic engineering applications due to their high sequence specificity and unidirectional integration reaction.
Structure
Serine integrases are made up of two conserved regions: an N-terminal catalytic domain (NTD) of 130 residues and a C-terminal domain of 300-500 residues (CTD). A long helix (E) connects the domains. The NTD contains the catalytic serine residue responsible for the enzyme family name and is also involved in attachment site synapsis (association), subunit rotation, and strand exchange. E helps to form the hydrophobic dimerization as well as tetramerization interfaces and also binds to the DNA minor groove near the attachment sites’ centers.
A prophage is a viral genome that integrates into the genome of a bacterium. In summary, it stays in the lysogenic cycle and is passed down through generations via genome replication. However, in order to enter the lytic cycle and destroy the bacterial cell, it can undergo excision from the host genome. Bacteriophages are bacteria-infecting viruses.
[…] or foes? The role of prophages in environmental adaptation and virulence of pectinolytic bacteria: Dickeya spp. and Pectobacterium […]
[…] Pride’s lab focuses on bacteriophage and human virome/microbiome analysis on the UCSD main campus. Clinical research studies are […]