In 21st-century research, where sequences burgeon, the demand for efficient and scalable genomic annotation tools has never been more pronounced. The recent arrival of Pharokka, an innovative annotation tool, has sparked considerable interest as it promises a seamless, rapid, and consistent approach to unraveling phage genomes.
Existing tools like RAST, PHASTER, and CPT Galaxy, though valuable, are web-server dependent and might pose challenges, especially when annotating numerous phage genomes in succession. Pharokka addresses this concern, embracing the one-line approach similar to the well-received Prokka. This simplicity is further enhanced by its tailored focus on phage genomes, a unique selling point that sets it apart from its predecessors.
A One-Line Marvel
The beauty of Pharokka lies in its accessibility and adaptability. It readily accepts DNA sequences in FASTA format, accommodating diverse scenarios. Whether you’re dealing with single complete contigs, incomplete assemblies, or even multiFASTA formats for metagenomic samples, Pharokka seamlessly steps into the fray. Its design even caters to metagenomically assembled phage genomes and genomic contigs, extending its utility and appeal.

Feature Prediction: Tailoring to Phages
Pharokka’s prowess is embodied in its strategic choice of feature prediction tools. By default, it employs PHANOTATE, a gem tailored for predicting coding sequences (CDS) in phage genomes. This selection is well-justified, as PHANOTATE excels in deciphering unique phage genome characteristics, like compact gene sizes and alternate start codons. Alternatively, users can opt for Prodigal, a versatile gene predictor perfect for large metavirome datasets.
Functional Gene Annotation
The heart of Pharokka’s potency lies in its functional gene annotation. Pharokka harnesses the power of the PHROGs database, home to a staggering 38,880 protein orthologous groups, each with a designated functional category. Predicted CDS are ingeniously aligned with the PHROGs database using mmseqs2, painting a vivid functional picture of each gene.
Beyond the Basics: Virulence and Resistance
Pharokka doesn’t stop at gene prediction and functional annotation. It takes a step further by detecting virulence factors and antimicrobial resistance genes. This facet, crucial for phage therapy applications, integrates the Comprehensive Antibiotic Resistance Database (CARD) and the Virulence Factor Database (VFDB), ensuring a comprehensive assessment.
Output: A Treasure Trove
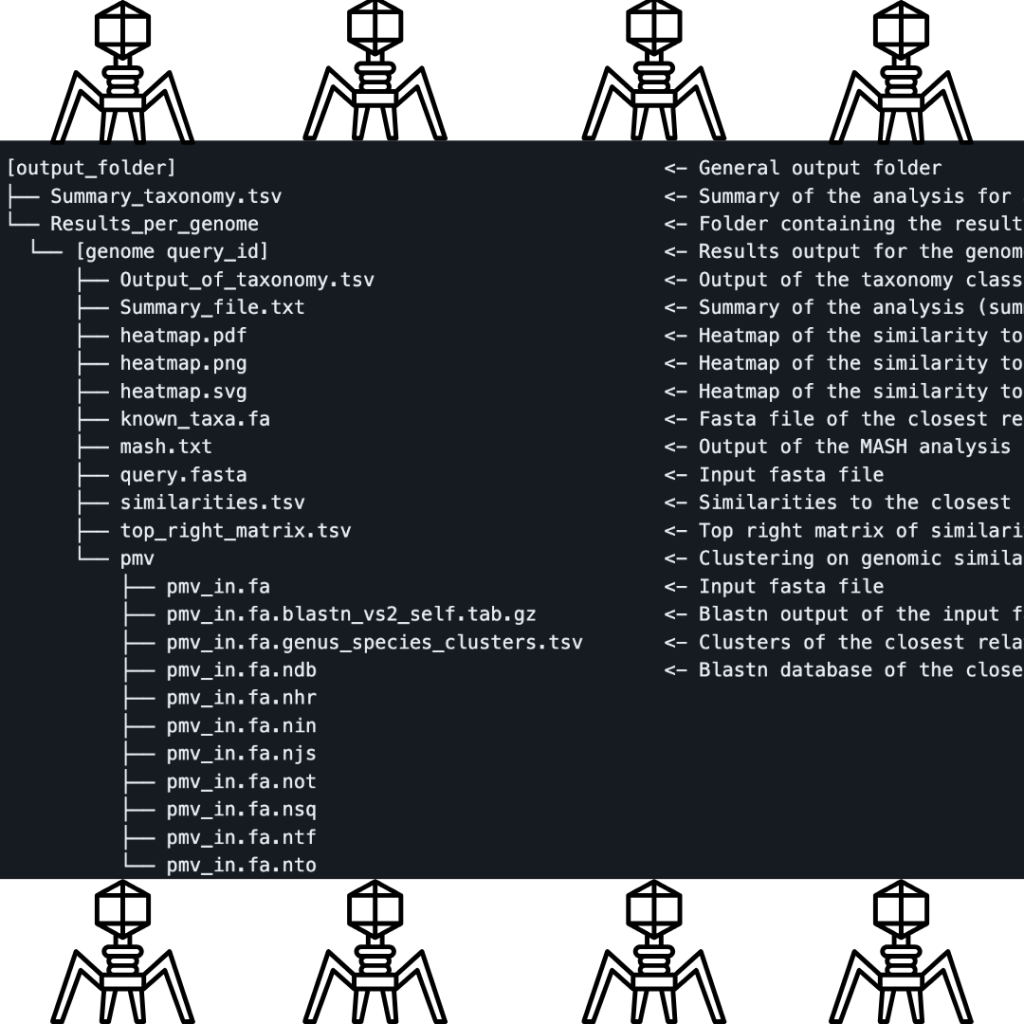
Pharokka’s output encompasses a rich array of files that even someone with partial familiarity with bioinformatics can grasp. The primary .gff file opens doors to downstream pan-genome endeavors. Other files include .tbl files for NCBI integration, a cds_functions.tsv file for insightful counts, and a length_gc_cds_density.tsv file detailing contig specifics. As the curtain closes, Pharokka’s contig-level summary unveils unique features in metaviromes, offering a window into potential stop codon reassignments or intriguing genetic landscapes.
A New Era in Phage Annotation
In the evolving landscape of phage genomics, Pharokka emerges as a potent ally. Its speed, simplicity, and tailored focus on phages set it on a pedestal. As phage researchers embark on the journey of genomic unraveling, Pharokka promises a smoother ride, simplifying the annotation process while revealing the rich functional tapestry of these elusive entities.
From my perspective, I believe that even if you’re not well-versed in command-line bioinformatics, it’s still worthwhile to consider trying out this tool, especially if you’re dealing with a substantial number of sequences requiring annotation, given its user-friendly nature. However, if you find yourself needing to annotate just one or two phage genomes, you might want to explore web-based tools, as highlighted in the introduction section. I’ve personally utilized a few of these web-based tools, and they’ve proven effective, although they might offer slightly less flexibility in tailoring the output to your preferences when compared to terminal-based tools like PHAROKKA.
For more information about PHAROKKA visit this GitHub page and publication George Bouras, Roshan Nepal, Ghais Houtak, Alkis James Psaltis, Peter-John Wormald, Sarah Vreugde, Pharokka: a fast scalable bacteriophage annotation tool, Bioinformatics, Volume 39, Issue 1, January 2023, btac776, https://doi.org/10.1093/bioinformatics/btac776. To read about many other amazing tools on our website please visit the bioinformatic tools category and our exclusive listing page.


Usually I do not read article on blogs, however I would like to say that this write-up very compelled me to take a look at and do so! Your writing taste has been amazed me. Thanks, quite nice post.