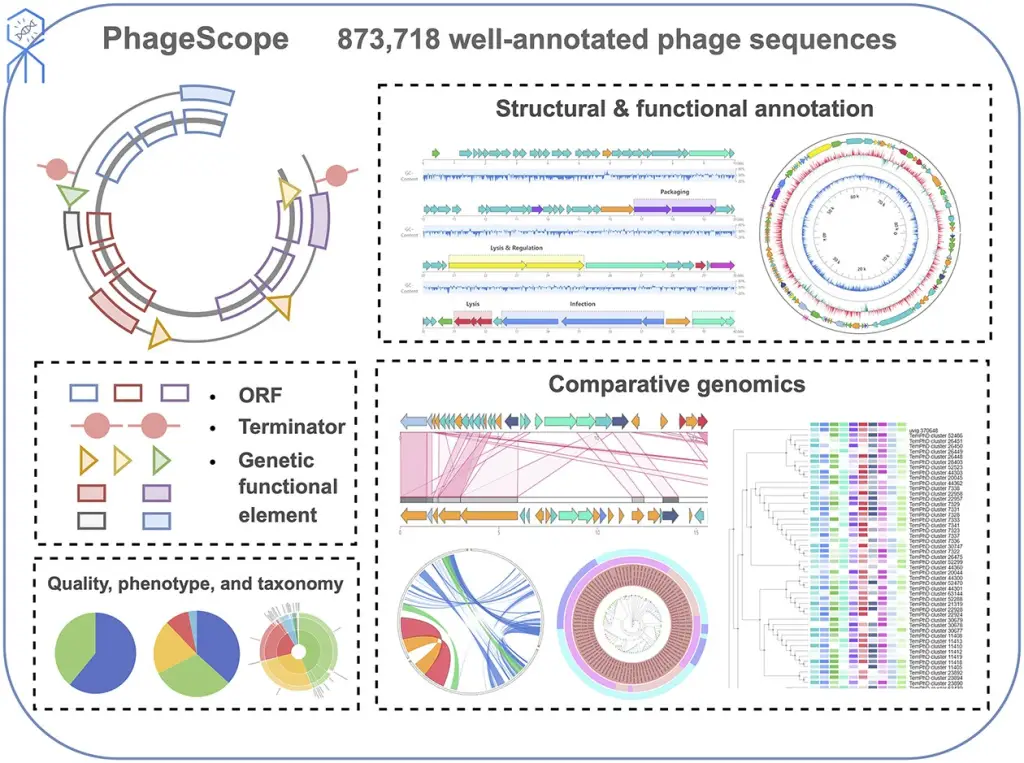
In the dynamic realm of bioinformatics, a transformative tool has emerged to unravel the intricacies of bacteriophages. PhageScope, a cutting-edge web-based tool developed by scientists from City University of Hong Kong, beckons researchers into the fascinating world of phage genomics. In this in-depth exploration, we delve into the nuances of PhageScope’s methodology, applaud its novel features, and scrutinize its performance, all through the lens of an enthusiast.
The Heart of PhageScope’s Power
At the core of PhageScope lies a colossal dataset comprising 873,718 phage sequences meticulously curated from public repositories and datasets, a testament to the tool’s commitment to exhaustiveness. The journey begins with an exhaustive search across RefSeq, Genbank, EMBL, and DDBJ, coupled with keyword mining and the integration of diverse datasets like PhagesDB and IMG/VR. This meticulous curation culminates in a dataset ripe for exploration.
The Capabilities of PhageScope
Genome Annotation
- Completeness Assessment
- Phenotype Annotation
- Host Assignment
- Lifestyle Prediction
- Structural Annotation
- ORF Prediction & Protein Classification
- Transcription Terminator Annotation
- Taxonomic Annotation
- Functional Annotation
- tRNA & tmRNA Gene Annotation
- Anti-CRISPR Protein Annotation
- CRISPR Array Annotation
- Virulent Factor & Antimicrobial Resistance Gene Detection
- Transmembrane Protein Annotation
Genome Comparison
- Sequence Clustering
- Sequence Alignment
- Comparative Tree Construction
Genome Annotation
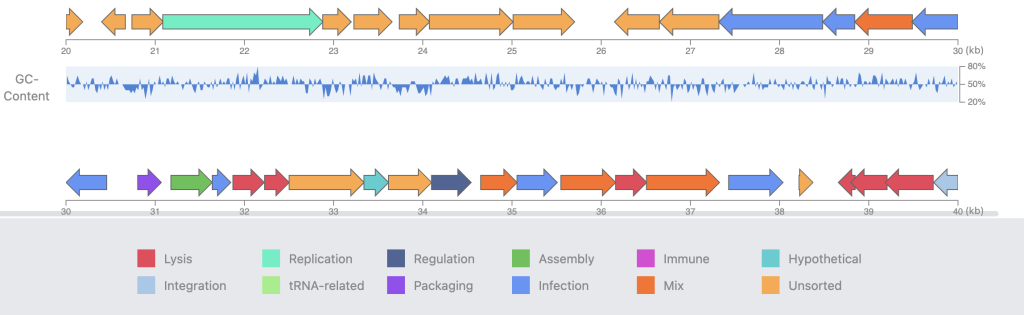
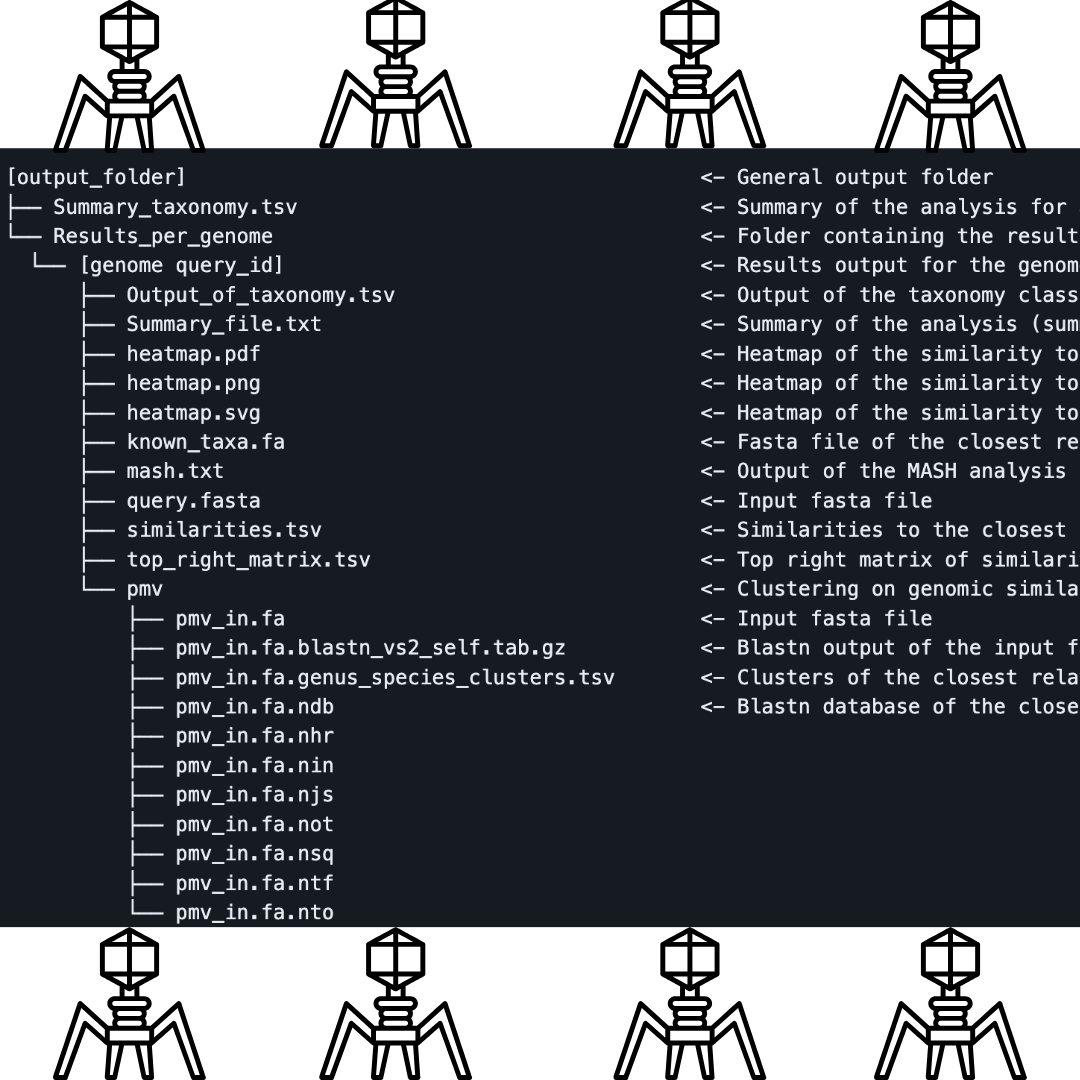
PhageScope’s strength lies not just in the dataset but in its application of state-of-the-art tools for systematic genome annotations. The completeness assessment, a critical step, employs CheckV v0.8.1 to categorize genomes into tiers, ensuring users navigate a landscape of reliable data. The phenotype annotation, a nuanced dance between DeepHost and homology searches, not only assigns host taxonomies but also predicts the elusive phage lifestyles. Structural annotation, a complex orchestration of Prodigal, Eggnog-mapper, and TransTermHP, brings to light the coding regions, terminators, and functional classes of proteins.
Taxonomy Assignment
To assign taxonomy, PhageScope employ HMMs to align phage proteins against taxonomically representative VOGs. This meticulous taxonomic annotation ensures precise classification, an essential component in the intricate dance of genomic exploration.
Functional Annotation
Functional annotation, a crucial aspect of any genomic study, is where PhageScope truly shines. The tool employs tRNAscan-SE, ARAGORN, AcRanker, and CRISPRCasFinder, among others, to identify tRNA and tmRNA genes, anti-CRISPR proteins, and CRISPR arrays. The integration of mmseqs for homology searches against VFDB and CARD completes the picture, revealing virulence factors and antimicrobial resistance genes, if the matches meet stringent thresholds.
Comparative Genomics
The tool’s prowess in comparative genomics is evident in its sequence clustering, sequence alignment, and comparative tree construction. Mmseqs takes center stage, generating subclusters and clusters with representative sequences. BLASTP performs pairwise alignments, showcasing coverage and identity values, while Alfpy and the neighbor-joining algorithm construct a comparative tree, unveiling hierarchies among multiple phages.
User Experience (The phage experience)
My hands-on exploration of PhageScope uncovered a user-friendly platform that marries functionality with aesthetics. The platform has a well designed page with easy to use functionality, having a range of tasks that a well sectioned. What I particularly appreciate about this tool is the availability of sample data, referred to by the developers as demo data. These datasets not only allow for visualization but also provide the option to execute, offering a practical insight into potential outcomes before processing one’s own samples. The inclusion of demo data is a time-saving feature, enabling users to familiarize themselves with the tool and anticipate real results without the extended wait times associated with processing actual data. The shortened run time for samples further enhances the user experience, ensuring efficient exploration and understanding of the tool’s capabilities.
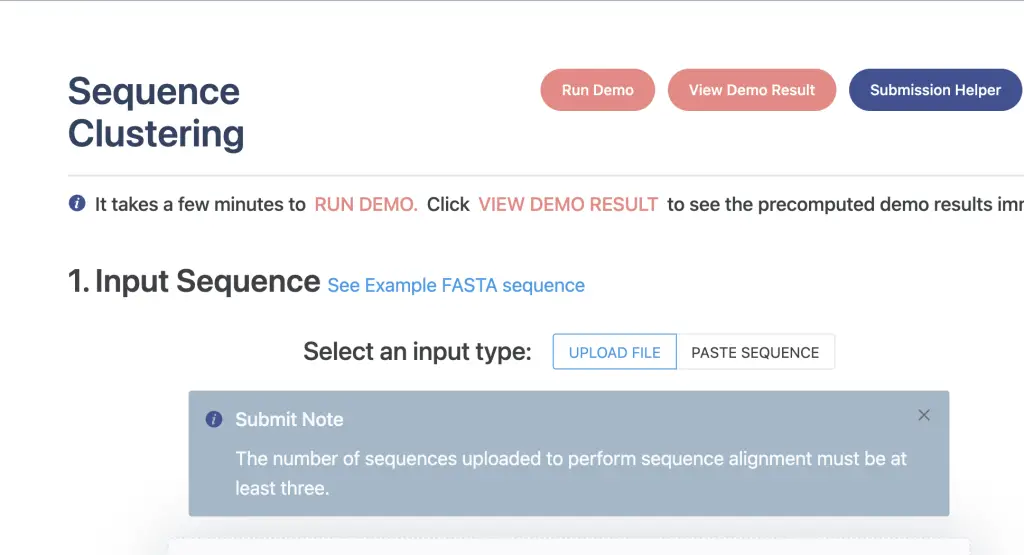
An added advantage of the tool is that the developers have made it freely accessible without the need for user accounts. The work runs are automatically saved, and the results are easily accessible (downloadable), providing a seamless and user-friendly experience.
While the tool provides preset color contrasts and the option to view genomes in both linear and circular formats, certain limitations have become apparent. Specifically, the tool lacks fine controls for data manipulation. Additionally, the limited database led to inaccurate host predictions for certain phages associated with rare bacteria. Furthermore, intermittent delays suggested potential server strain during peak usage times.
PhageScope’s Future
PhageScope is not just a tool; it’s an expedition into the unexplored territories of bacteriophage genomics. As the scientific community eagerly anticipates updates and refinements, PhageScope remains poised to leave an indelible mark on the trajectory of phage biology and microbial ecology studies. The developers have done a very outstanding work on bringing this tool to life. I can surely recomend it for someone who has would like to use web based interface to annotate their phage. Also it can suite someone who want to get nice photos for their phage genomes. You can access PhageScope here. For more tools, please visit our tools section by clicking here
Reference
The cover image has been sourced from the original published study published by Ruo Han Wang, Shuo Yang, Zhixuan Liu, Yuanzheng Zhang, Xueying Wang, Zixin Xu, Jianping Wang, Shuai Cheng Li, PhageScope: a well-annotated bacteriophage database with automatic analyses and visualizations, Nucleic Acids Research, 2023;, gkad979, https://doi.org/10.1093/nar/gkad979
Abbreviations
| Abbreviation | Full Form | Explanation for Layman |
|---|---|---|
| AcRanker | Anti-CRISPR prediction tool | A tool that predicts the presence of anti-CRISPR proteins, which can counteract CRISPR-based bacterial defenses. |
| Alfpy | Alignment-Free Sequence Comparison Method | A method for comparing genetic sequences without aligning them, aiding in efficient analysis. |
| BLASTP | Basic Local Alignment Search Tool for Proteins | A tool that searches for similar protein sequences in databases, helping identify proteins with shared features. |
| CARD | Comprehensive Antibiotic Resistance Database | A database that compiles information on antibiotic resistance genes, aiding in the study of drug resistance. |
| CheckV | Quality Assessment Tool for Metagenome-Assembled Genomes | A tool that assesses the quality of genomic data assembled from microbial communities. |
| CRISPRCasFinder | CRISPR array detection tool | A tool that identifies CRISPR arrays, which are part of a bacterial defense mechanism against viruses. |
| DDBJ | DNA Data Bank of Japan | A database storing DNA sequences, serving as a valuable resource for genetic research. |
| DeepHost | Host Prediction Tool using Deep Learning | A tool that uses deep learning algorithms to predict the hosts (bacteria) that phages infect. |
| Eggnog-mapper | Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups | A tool that assigns evolutionary relationships to genes based on shared functions. |
| EMBL | European Molecular Biology Laboratory | A leading research institute supporting molecular biology studies and providing valuable data resources. |
| GPD | Global Phage Database | A worldwide database collecting information on bacteriophages, viruses that infect bacteria. |
| GOV2 | Global Ocean Virome 2 | A global initiative studying viruses in the ocean, contributing to our understanding of marine ecosystems. |
| GVD | Global Virome Database | A global database cataloging viral diversity, enhancing our knowledge of viruses and their impact. |
| IMG/VR | Integrated Microbial Genomes with Viral Resources | A platform integrating microbial and viral genome data for comprehensive analysis. |
| IGVD | Integrated Global Viral Database | A global database consolidating viral information, facilitating research on global viral diversity. |
| MGV | MetaGenomic Viral Database | A database focused on viruses identified through metagenomic studies, aiding in viral genome analysis. |
| PHROG | Phage and Host Relations Ontology Graph | A tool that helps researchers understand the relationships between bacteriophages and their host bacteria. |
| Prodigal | Prokaryotic Dynamic Programming Genefinding Algorithm | A tool that identifies genes in the DNA of bacteria, assisting in understanding bacterial genetic information. |
| RefSeq | Reference Sequence Database | A comprehensive database providing reference DNA sequences for various organisms, supporting genetic research. |
| STV | Siphoviridae Viral Database | A database specializing in a family of bacteriophages called Siphoviridae, aiding research on these viruses. |
| TemPhD | Temperate Phage Database | A database focused on temperate phages, which can integrate into bacterial DNA and remain dormant. |
| TMHMM | Transmembrane Helix Prediction Tool | A tool that predicts the presence of transmembrane helices in proteins, aiding in the study of protein structure. |
| TransTermHP | Transcriptional Terminator Prediction for Bacterial Genomes | A tool that predicts termination signals in bacterial DNA, assisting in understanding gene regulation. |
| tRNAscan-SE | tRNA detection in genomes | A tool that identifies transfer RNA (tRNA) genes in genomes, essential for protein synthesis. |
| VFDB | Virulence Factor Database | A database containing information on virulence factors, which contribute to the severity of infections. |
| VOG | Viral Orthologous Groups | A system that groups together genes from different viruses that share evolutionary ancestry. |

Leave a Reply
You must be logged in to post a comment.