Prophages are integrated viral genomes that reside within bacterial chromosomes, remaining dormant until triggered by various environmental stimuli. One of the most widely employed agents for prophage induction is Mitomycin C, a DNA cross-linking agent known for its ability to induce DNA damage. This article explores the molecular mechanisms behind prophage induction using Mitomycin C, shedding light on its applications in the study of bacteriophages and their impact on bacterial populations.
Prophages, once considered silent passengers within bacterial genomes, have emerged as essential players in bacterial evolution, contributing to genetic diversity and adaptation. Prophage induction, the process by which these latent viral genomes are activated and enter the lytic cycle, is a crucial phenomenon with broad implications for bacterial physiology and ecology. Among the various methods employed for prophage induction, Mitomycin C has proven to be a powerful tool due to its ability to induce DNA damage.
Mitomycin C and DNA Damage:
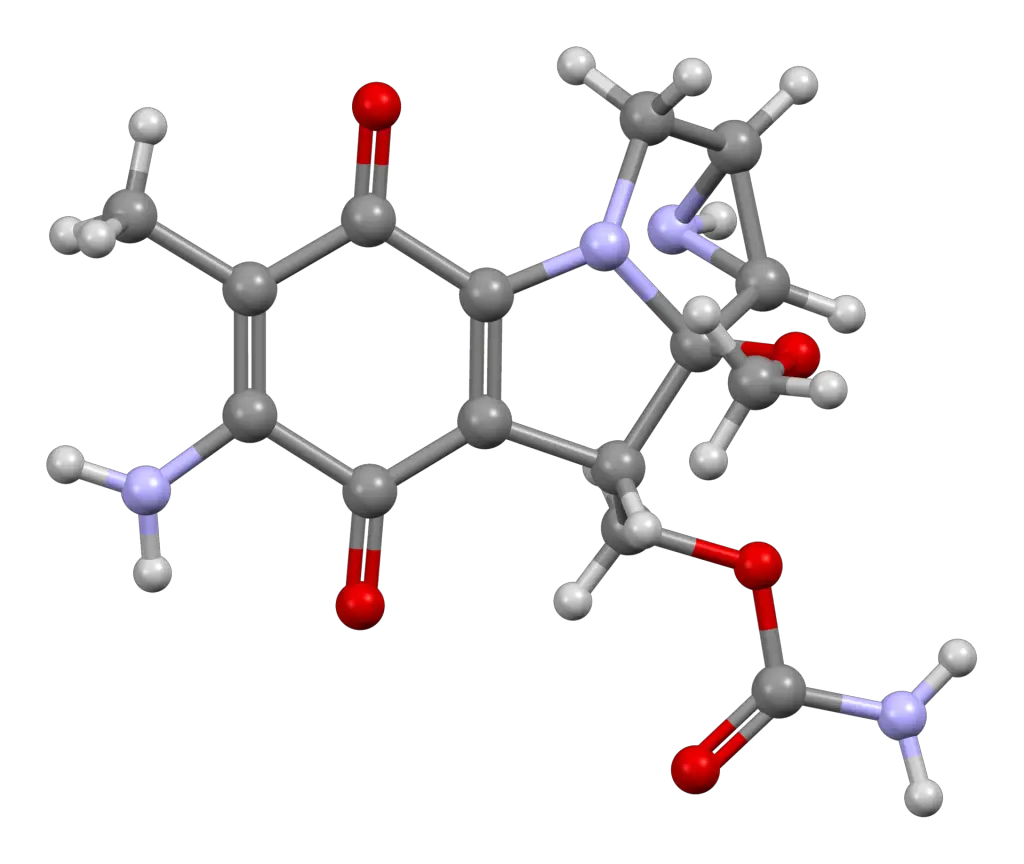
Mitomycin C, a natural antibiotic produced by Streptomyces caespitosus, is renowned for its DNA cross-linking properties. It forms covalent bonds between adjacent guanine bases, resulting in interstrand cross-links that impede DNA replication and transcription. The propensity of Mitomycin C to induce double-strand breaks and DNA damage triggers the SOS response in bacteria, a key regulatory network involved in DNA repair and mutagenesis.
SOS Response and Prophage Induction:
The SOS response acts as a sentinel mechanism, sensing DNA damage and orchestrating a cascade of events to mitigate cellular stress. Central to this response is the activation of the RecA protein, which, upon binding to single-stranded DNA exposed during DNA damage, becomes a catalyst for the autocleavage of the LexA repressor. The derepression of LexA-regulated genes, including those responsible for prophage excision and induction, facilitates the transition from lysogeny to the lytic cycle.
Protocol: Prophage Induction Using Mitomycin C
Note: The following protocol provides a step-by-step guide for performing prophage induction using Mitomycin C in a bacterial culture. Ensure proper safety measures, and adhere to institutional guidelines for handling chemicals and biological materials.
Materials:
- Bacterial culture containing the target prophage
- Mitomycin C (Sigma-Aldrich or equivalent)
- LB broth or appropriate bacterial growth medium (Read detailed step by step procedure on preparation of LB agar here)
- Agar plates for bacterial culture and plaque assay
- Incubator
- Centrifuge and centrifuge tubes
- Spectrophotometer or OD600 meter
- Sterile pipettes and tips
- Water bath (This can be employed to maintain the overlay agar at an optimal temperature)
Procedure:
1. Preparation of Bacterial Culture:
a. Inoculate a single colony of the bacterial strain containing the target prophage into a fresh LB broth or any other suitable growth medium.
b. Incubate the culture overnight at 37°C with shaking (200 rpm) until reaching mid-log phase (OD600 ~ 0.4-0.6).
2. Mitomycin C Treatment:
a. Calculate the appropriate concentration of Mitomycin C based on previous literature or pilot experiments (the concentration may be host dependant). Typically, concentrations range from 0.1 to 1.0 μg/ml some studies have gone up to 3 μg/ml.
b. Add Mitomycin C to the bacterial culture to achieve the desired concentration. Mix gently by swirling or pipetting.
c. Incubate the culture with Mitomycin C at 37°C (incubation conditions depend on the host in question) with shaking for a predetermined duration (commonly 2-4 hours) to allow sufficient time for prophage induction. For slow-growing bacteria, extended incubation periods may be necessary to allow for sufficient induction.
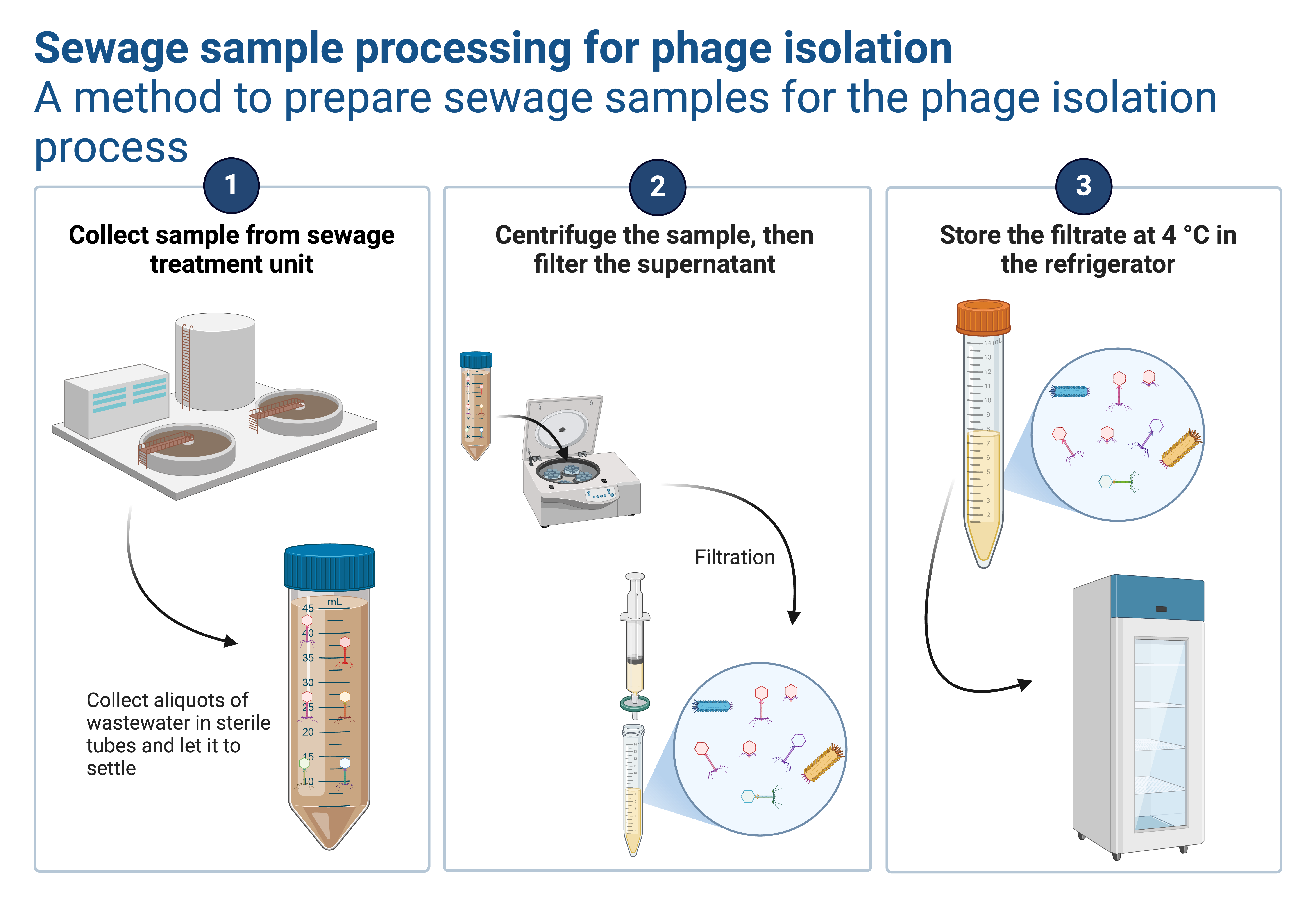
3. Harvesting induced prophages:
a. After the incubation period, sediment the bacterial cells from the sunspension by centrifugation at 4000 rpm for 10-15 minutes at room temperature.
b. Discard the the pellet (these are bacterial cells) and use the supernatant for next stage (virions will be suspended in the supernatant since they are lighter).
4. Filtering the induced prophages:
a. Use a 0.22 μm syringe filter or any other favourable filter of the same size to filter the supernatant in to clean and sterile tube (Read about filtration from this article). This process will supplement centrifugation in eliminating any residual bacterial cell debris that could persist in the supernatant..
b. Keep the your filtrate at the refrigeration temperature (2-8 oC) ready for confirming if your induction was successfull.
5. Confirming Prophage Induction:
a. By performing plaque assay and observing the formation of plaques on you plate. Despite this approach being optimal many cases, some scientists have reported prophages reintegrating instead of producing plaques. There fore, negative results at this stage might prompt the consideration of alternative methods. In such cases, you may need to explore one of the following (option b,c or d) alternative approaches.
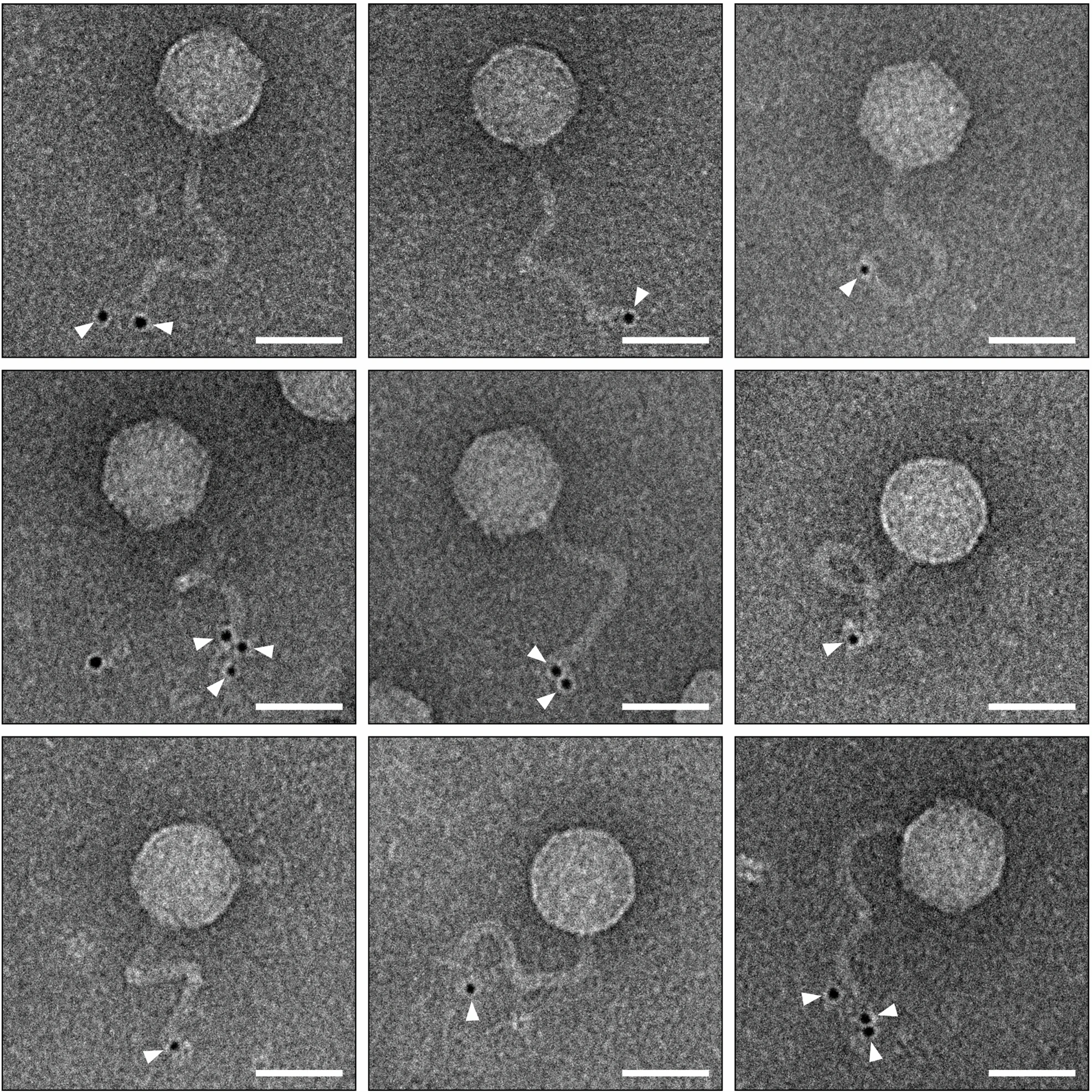
b. Performing transmission electron microscopy to visualize phage particles in your filtrate (lysate). This will deeply depend on the availability of the facility.
c. Confirm prophage induction by isolating phage DNA from the induced culture using standard phage DNA extraction protocols. It’s crucial to consider that host DNA might also be present, and its size could allow it to pass through the filter pores. Therefore, it may be necessary to incorporate a nuclease step to digest free-floating nucleic acids before proceeding with the extraction of phage DNA. This precaution ensures a more precise and uncontaminated isolation of the desired genetic material.
d. Perform PCR targeting prophage-specific genes or regions to verify the presence and activation of the prophage.
Notes:
- Conduct all procedures in a sterile environment to avoid contamination..
- Adjust Mitomycin C concentration and incubation time based on the specific characteristics of the bacterial strain and prophage under investigation.
- Regularly monitor bacterial growth during the induction process to optimize the timing for prophage induction.
This protocol serves as a general guideline, and researchers are encouraged to tailor the procedure according to the specific requirements of their experimental setup and bacterial systems.
The phage blog
Molecular Insights into Prophage Induction:
The molecular events leading to prophage induction following Mitomycin C treatment involve a delicate interplay between host and viral factors. The RecA-mediated cleavage of the cI repressor, encoded by prophage repressor genes, liberates the phage genome, allowing for the expression of lytic genes and subsequent replication of phage DNA. Concurrently, the host cell experiences DNA damage repair processes, often leading to mutations and genomic alterations that shape bacterial evolution.
Applications and Significance:
Understanding the mechanisms of prophage induction using Mitomycin C has broad applications in diverse fields. In experimental settings, this approach enables the controlled induction of prophages for the isolation and study of bacteriophages. Moreover, it provides insights into the ecological dynamics of bacterial populations, as prophage induction influences bacterial virulence, pathogenicity, and adaptability. The study of Mitomycin C-induced prophage induction has practical implications in biotechnology, where engineered bacteriophages are employed for various applications, including phage therapy and bacterial biocontrol.
To access additional protocols, please click here. Don’t miss out on intriguing articles like this – subscribe to our newsletter today! Stay updated on the latest scientific insights, research findings, and valuable content delivered straight to your inbox. Subscribe now for a continuous stream of engaging and informative material. Also you can follow the phage on X(formally twitter), Facebook, and LinkedIn page
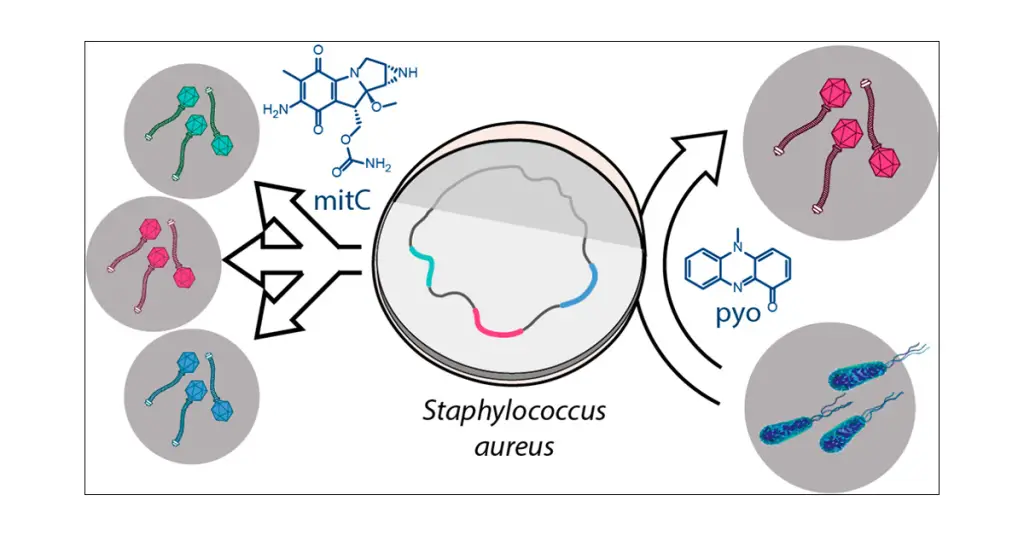
Cover photo credit: Jancheva and Bottcher, A Metabolite of Pseudomonas Triggers Prophage-Selective Lysogenic to Lytic Conversion in Staphylococcus aureus. 2021.

Leave a Reply
You must be logged in to post a comment.