A groundbreaking study from the Department of Energy’s Lawrence Berkeley National Laboratory. This research introduces an innovative method to swiftly label and characterize bacteriophages, viruses that target bacteria, using the acclaimed CRISPR gene-editing technology. The team’s approach holds the promise of transforming our ability to identify, track, and manipulate these elusive entities.
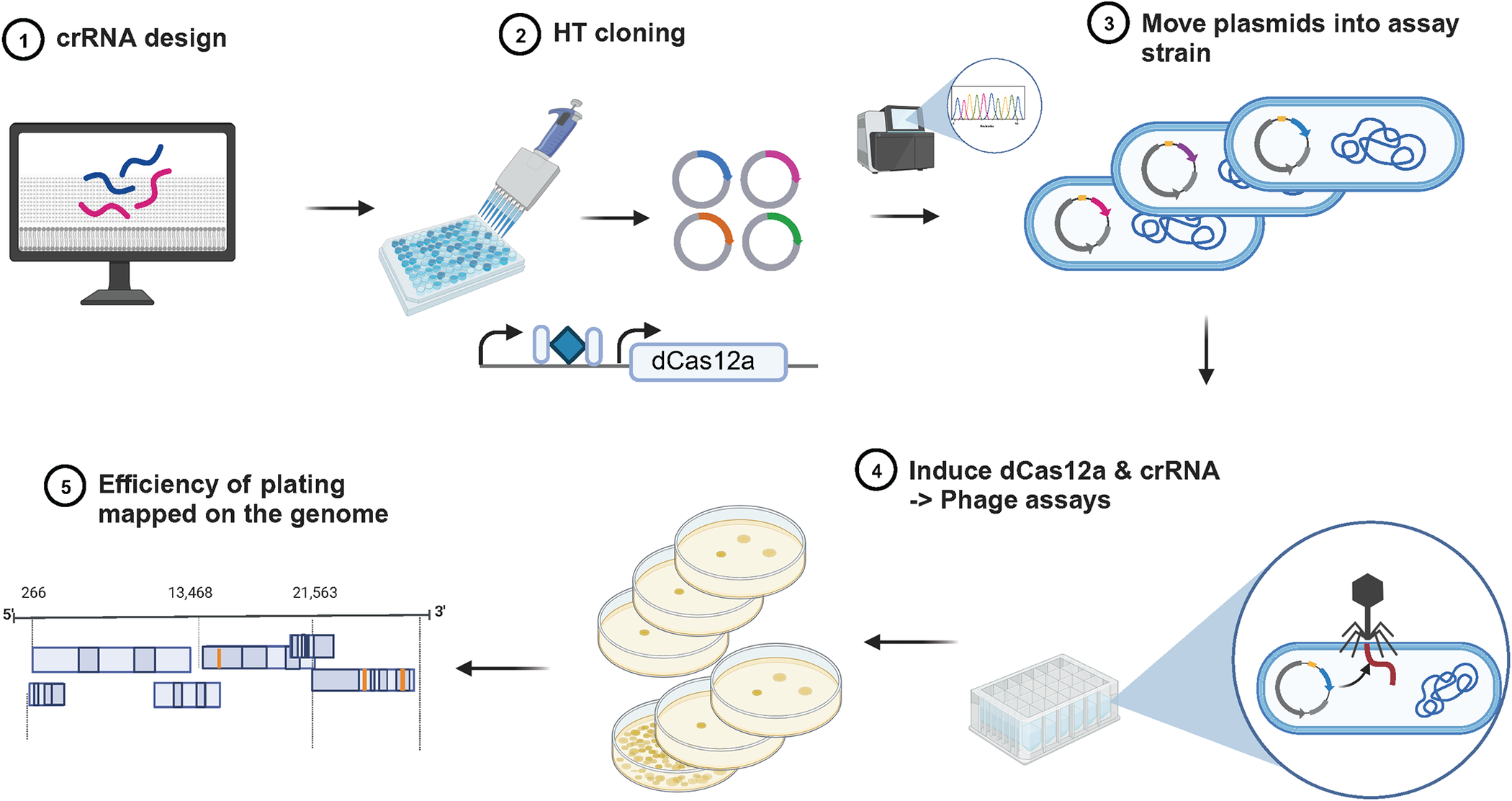
Central to the researchers’ methodology is the mapping of the gene essentiality landscape of phages on a genome-wide scale. Employing CRISPR interference (CRISPRi) technology, the team systematically suppressed the expression of nearly every gene in two model phages, λ and P1. This groundbreaking effort marks the first systematic study of gene essentiality at such a scale in bacterial viruses.
By discerning which genes are indispensable for the infection cycle and which are dispensable, the researchers provide a comprehensive understanding of phage gene essentiality. Crucially, this knowledge extends beyond specific phage types, highlighting an interconnectedness among phage genes that could unlock further insights into their functioning.
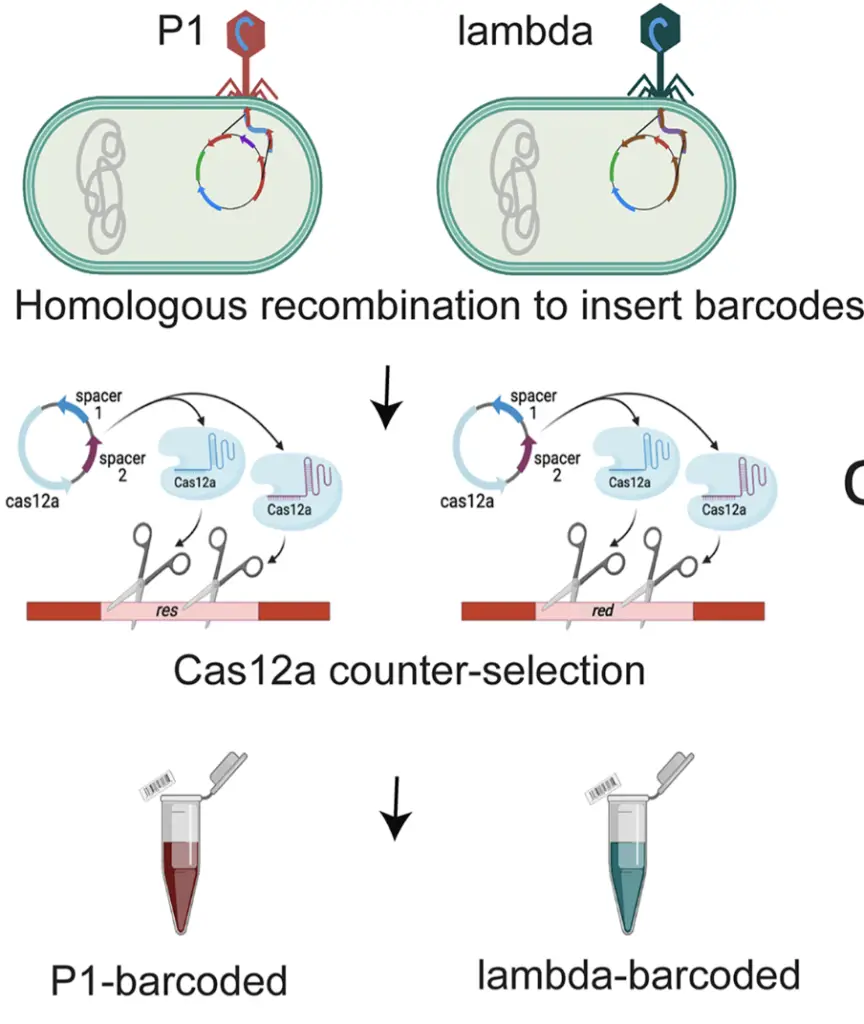
By putting certain genes on mute, the researchers stumbled upon a game-changing revelation in their study—the incorporation of barcodes into phage genomes. The objective was to pinpoint genes that weren’t crucial for phage function and swap them with distinctive tags. These tags, comprising unique sequences of nucleic acids, operate much like the barcodes you find at your local supermarket. This ingenious move now allows scientists and clinicians to swiftly pinpoint and monitor various phages in a range of environments. This, in turn, equips researchers with a powerful tool to delve into diverse aspects of bacteriophages within a specific setting, opening up new avenues for scientific exploration.
Bacteriophages represent one of the most abundant and genetically diverse entities. By combining the power of CRISPR gene-editing technology with the practicality of barcoding, the researchers have opened new avenues for understanding, utilizing, and tracking bacteriophages. This pioneering work not only sheds light on the gene essentiality of phages but also paves the way for transformative applications across agriculture, environmental science, and healthcare. As we delve deeper into the microbial universe, the potential unleashed by this study offers a glimpse into a future where the untapped knowledge of phage gene function finds practical and impactful applications.
You can read their full paper here Denish Piya et al, Systematic and scalable genome-wide essentiality mapping to identify nonessential genes in phages, PLOS Biology (2023). DOI: 10.1371/journal.pbio.3002416. Cover photo credit to the original study
Have you liked what you have just read, click here to read more related content (phage-related news) like this one and also join our community by following the phage on X(formally Twitter), Facebook, and LinkedIn page