Bacteriophages have gained general popularity in recent years due to their potential in treating multidrug-resistant bacteria. Although they may seem new to scientists in the medical field, that is not true in the biotechnology field, as scientists have been routinely using them for decades. Through phage display, scientists have been studying and manipulating proteins in laboratories. Since its development in the mid-1980s by Nobel prize winner G. P. Smith, this technology has enabled the identification of molecules with high affinity and specificity for their targets, driving significant advancements in diagnostics, therapeutics, and basic research.
What is Phage Display?
Phage display is a molecular biology technique that involves manipulating phage nucleic acid by adding a foreign DNA sequence which will be expressed as a peptide on the surface of a phage. These foreign peptides are displayed as fusions with the protein coat of the virus. This method enables the screening and identification of molecules that bind with high specificity to various targets. While various phages can be employed in this method, the most commonly used system has historically been filamentous phages such as M13. These (filamentous phages) are now classified under Monodnaviria › Loebvirae › Hofneiviricota › Faserviricetes › Tubulavirales › Inoviridae.
Since phages are viruses, they have several advantages that make them useful for display. Among them, their small genome makes it easy to manipulate and understand the function of each gene, making it simple to insert foreign genes. Additionally, scientists are aware of bacteriophages and their non-pathogenic nature, also they multiply quickly and are relatively stable particles.
The process typically involves the following steps:
You can also read this section on the protocol page: Phage display technique
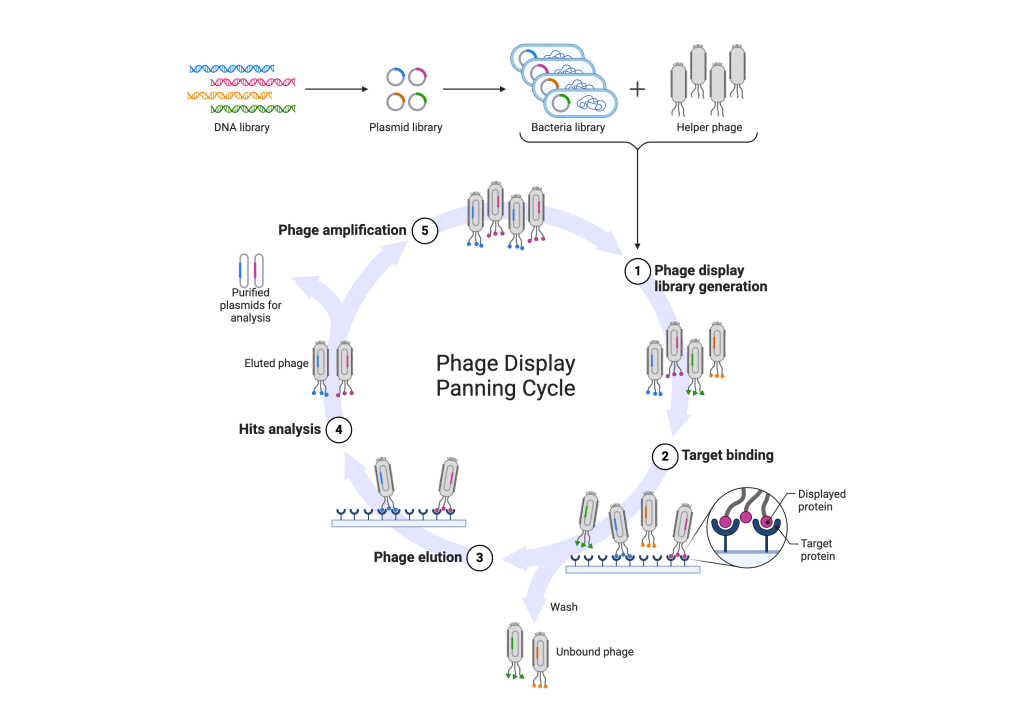
- Library Construction (Library Generation): In the initial phase, a diverse library of DNA sequences encoding peptides or proteins is generated. Each sequence represents a unique variant that could potentially bind to a target molecule. These sequences are then inserted into the genome of bacteriophages.
- Target Binding: The bacteriophages carrying the peptide or protein variants are exposed to the target molecule of interest. During this step, phages displaying peptides or proteins with binding affinity to the target molecule will selectively interact with it, forming specific complexes.
- Phage Elution: After binding to the target, the phage-target complexes undergo elution, usually through methods like pH alteration or the introduction of competitive agents. This step is designed to release the bound phages from the target molecule while preserving their integrity for subsequent analysis.
- Hits Analysis: The eluted phages, which contain potential binders to the target molecule, undergo analysis to identify successful interactions. This analysis may involve techniques such as sequencing, affinity assays, or functional assays to characterise the binding properties of the selected peptides or proteins.
- Phage Amplification: Phages demonstrating successful binding to the target molecule are amplified by infecting a suitable bacterial host. This amplification step enriches the phage population carrying the desired peptide or protein variants, thereby facilitating subsequent rounds of selection or downstream applications.
Applications of Phage Display
Phage display has a wide array of applications, especially in drug discovery, diagnostics, and basic research.
1. Antibody Discovery and Engineering
One of the most impactful applications of phage display lies in the discovery and engineering of monoclonal antibodies. Traditional methods, like hybridoma technology, can be time-consuming and labor-intensive. Phage display provides a swift and efficient alternative, enabling the production of antibodies with exceptional specificity and affinity for a diverse range of antigens.
- Therapeutic Antibodies: Numerous therapeutic antibodies developed through phage display have gained approval for clinical use, targeting diseases such as cancer, autoimmune disorders, and infectious diseases.
- Diagnostic Antibodies: Antibodies derived from phage display are utilised in various diagnostic assays, including ELISA and flow cytometry, to detect biomarkers associated with diseases.
2. Peptide Ligand Discovery
Phage display is also employed to identify peptide ligands that bind to specific targets, including proteins, nucleic acids, and small molecules. These peptides can serve various purposes such as:
- Inhibitors: Peptides that block the activity of target proteins, providing potential therapeutic agents for diseases where inhibition of specific protein functions is desirable.
- Molecular Probes: Peptides that bind to biomarkers can be used in imaging and diagnostic applications to visualise and detect disease states.
3. Protein-Protein Interaction Studies
Understanding protein-protein interactions is vital for deciphering cellular processes and pathways. Phage display can be utilised to map interaction sites and pinpoint critical residues involved in binding, thus offering insights into the molecular mechanisms underlying these interactions.
4. Vaccine Development
Phage display is utilised for identifying epitopes, which are specific parts of an antigen recognised by the immune system. By displaying these epitopes on the surface of phages, researchers can develop vaccines that provoke robust and targeted immune responses.
5. Synthetic Biology and Protein Engineering
Phage display enables the engineering of proteins with enhanced or novel functions which can be aided by insilico designing. Through screening large libraries of protein variants, researchers can select for properties such as increased stability, altered substrate specificity, or enhanced catalytic activity.
Advancements and Innovations in Phage Display
Since its inception, phage display technology has undergone numerous innovations that have expanded its utility and efficiency.
1. Next-Generation Sequencing (NGS)
The integration of NGS with phage display has significantly enhanced the speed, efficiency and depth of the screening step. This allows the rapid sequencing of millions of phage-displayed variants, providing a comprehensive view of binding interactions and enabling the identification of rare high-affinity binders.
2. High-Throughput Screening
Automation and robotic systems have been integrated into phage display workflows to enable precision by removing human error factors. This has accelerated the discovery process and allowed the simultaneous screening of multiple targets using less manpower.
3. Alternative Display Platforms
While filamentous bacteriophages such as M13 are commonly used in phage display, alternative platforms, including T4 and T7 bacteriophages, have been explored. These platforms offer different advantages, such as higher stability or the ability to display larger proteins.
4. Multivalent Display
Innovations in phage display have enabled the presentation of multiple copies of a peptide or protein on the same phage particle (multivalent display). This increases the avidity of interactions and can enhance the detection of low-affinity binders.
5. In Vivo Phage Display
In vivo, phage display involves the administration of phage libraries into living organisms to identify peptides or proteins that bind to specific tissues or cell types. This approach has been used to discover targeting ligands for drug delivery and imaging agents.
Challenges and Future Directions
Despite its many successes, phage display faces several challenges. The quality of the initial library, the efficiency of selection methods, and the risk of false positives can all influence the results. Moreover, displaying certain proteins on phages can be difficult if they need post-translational modifications that the bacterial host cannot provide.
Future directions for phage display research include:
- Improving Library Diversity: Improving the diversity and quality of phage display libraries enhances the likelihood of identifying high-affinity binders.
- Hybrid Approaches: Combining phage display with other technologies, such as CRISPR, enables the creation of more sophisticated screening platforms.
- Therapeutic Delivery: Developing phage-based delivery systems for therapeutic agents involves utilising phage display to target specific cells or tissues.
- Environmental and Agricultural Applications: Expanding the application of phage display to identify peptides and proteins for environmental monitoring, bioremediation, and agricultural biotechnology is crucial.
With its expanding capabilities, phage display is poised to continue playing a crucial role in molecular engineering and innovation, tackling significant challenges across industry, research, and healthcare.

Leave a Reply
You must be logged in to post a comment.