
Recently published research has made significant strides in understanding the complex interactions between bacteria and the viruses that infect them, known as phages. This discovery, published in Nature, sheds light on the mechanism phage protein uses to overcome a bacterial defense that was not known before.
The CRISPR-Cas system is by far the most well-known antiphage defense system extensively studied and understood by scientists, at least from the bacterial side. It is now widely utilized as a tool in biotechnology worldwide. However, when it comes to the mechanisms that phages use to counteract CRISPR, many are still partially or completely unknown, due to the previously underestimated impact of phages before the current surge of interest in phage therapy.
Bacteria use the CRISPR-Cas system to identify phage DNA and cleave it into fragments using specialized enzymes. Bacteriophages, in turn, have developed various methods to evade these actions by producing mechanisms involving proteins such as Chimalin coded by Jumbo phages. These systems render the CRISPR-Cas defense mechanism ineffective, akin to bacteria possessing a pair of scissors that do not function. Understanding these mechanisms provides a clearer picture of the ongoing microscopic arms race.
In the published article, researchers focused on anti-CRISPR-associated (Aca) protein that phages use to deploy anti-CRISPR mechanisms. Anti-CRISPR allows phages to block the CRISPR–Cas immune system of bacteria, a defense system bacteria utilize to protect themselves from viral infection. By understanding how phages counteract these defenses, scientists can identify appropriate phages to use as antimicrobials against bacterial pathogens in human health, agriculture and even beyond.
The research revealed that phages must carefully deploy their anti-CRISPR mechanisms. Aca protein, which contains a helix–turn–helix (HTH) domain, plays a crucial role in this process. The HTH domain is known for its ability to bind DNA sequences and regulate gene expression, either turning genes on or off. However, the study discovered that this domain is more versatile than previously understood. It not only binds DNA but also interacts with RNA transcripts, the molecules that mediate between DNA sequences and the anti-CRISPR encoded within them.
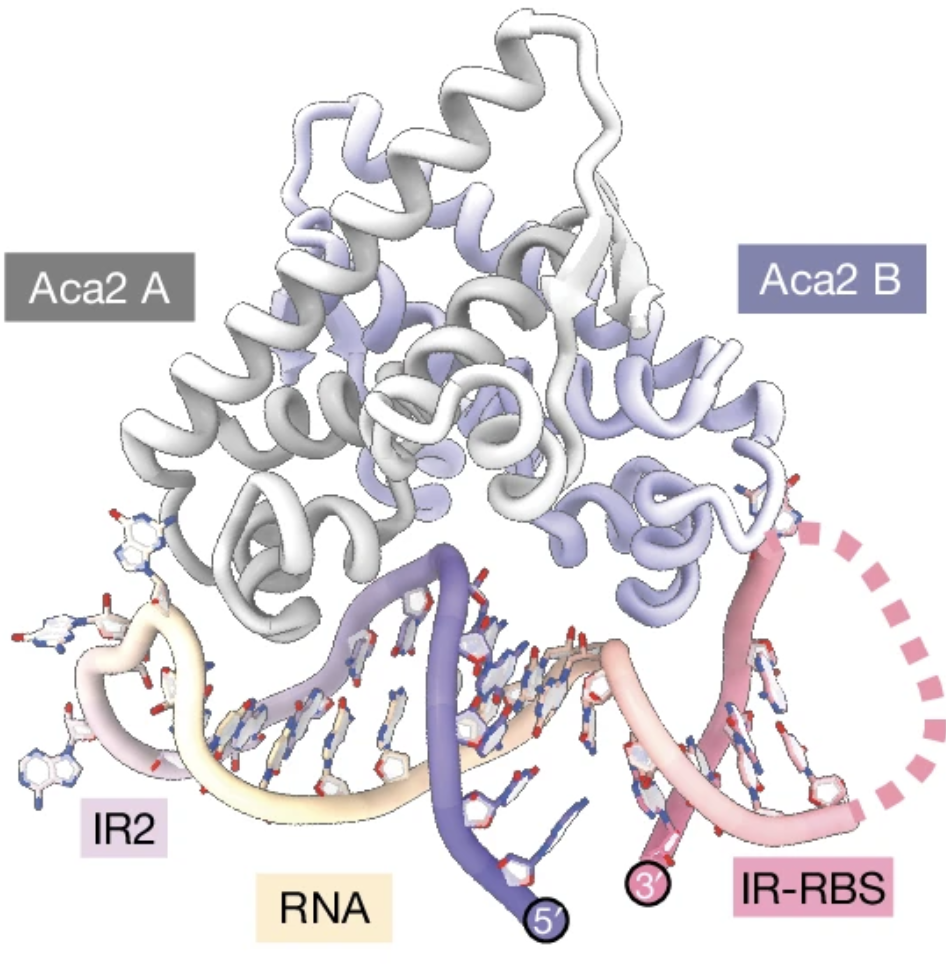
This dual functionality of the HTH domain means that it can regulate anti-CRISPR production at multiple levels. It controls gene expression through DNA binding and also inhibits the translation of mRNAs by binding to conserved RNA structures, preventing ribosome access. This layered regulation ensures that phages can rapidly replicate their DNA without causing toxic overexpression of anti-CRISPR genes, thereby maintaining a balance that is critical for their survival and effectiveness.
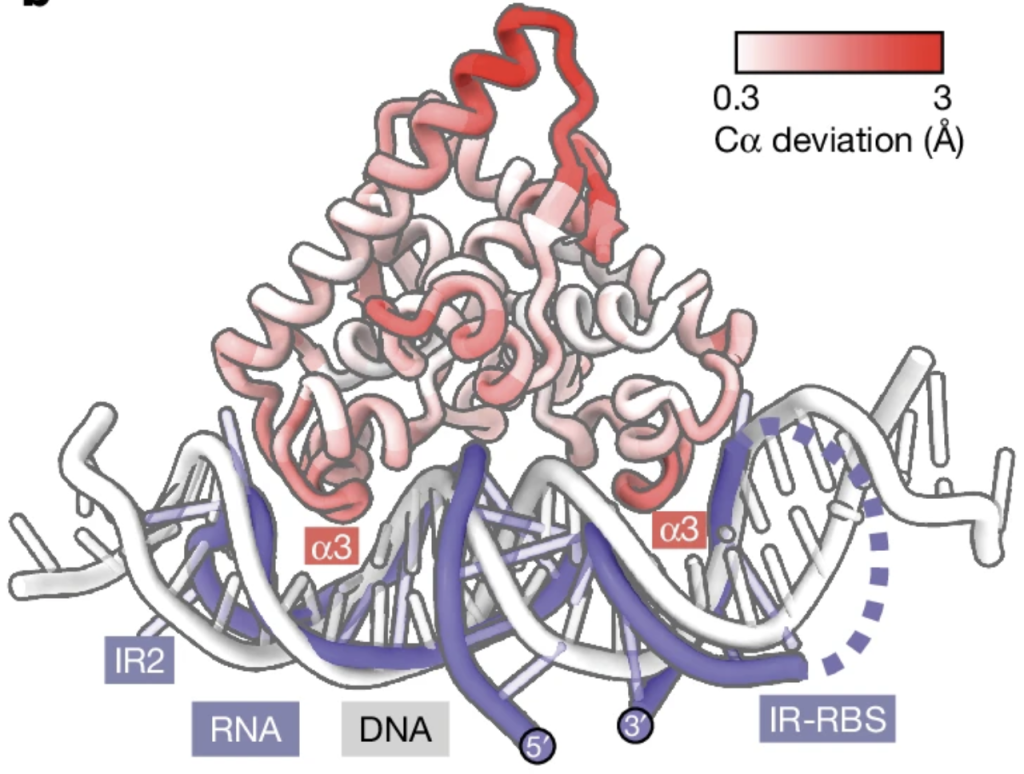
This discovery has significant implications for the scientific community’s understanding of anti-phage systems and gene regulation in phages. It came up with a novel regulatory mechanism within a well-studied family of proteins, potentially changing how researchers view the function and mechanism of these protein domains. By unravelling this complex regulation, the study advances our knowledge of how phages evade bacterial defenses and maybe like other phage characteristics, we may have a new method to use in biotechnology.
The study was published by Birkholz, N., Kamata, K., Feussner, M. et al. Phage anti-CRISPR control by an RNA- and DNA-binding helix–turn–helix protein. Nature (2024). https://doi.org/10.1038/s41586-024-07644-1 and MMDB citation Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, Bryant SH. " MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014 Jan; 42(Database issue):D297-303
Leave a Reply
You must be logged in to post a comment.