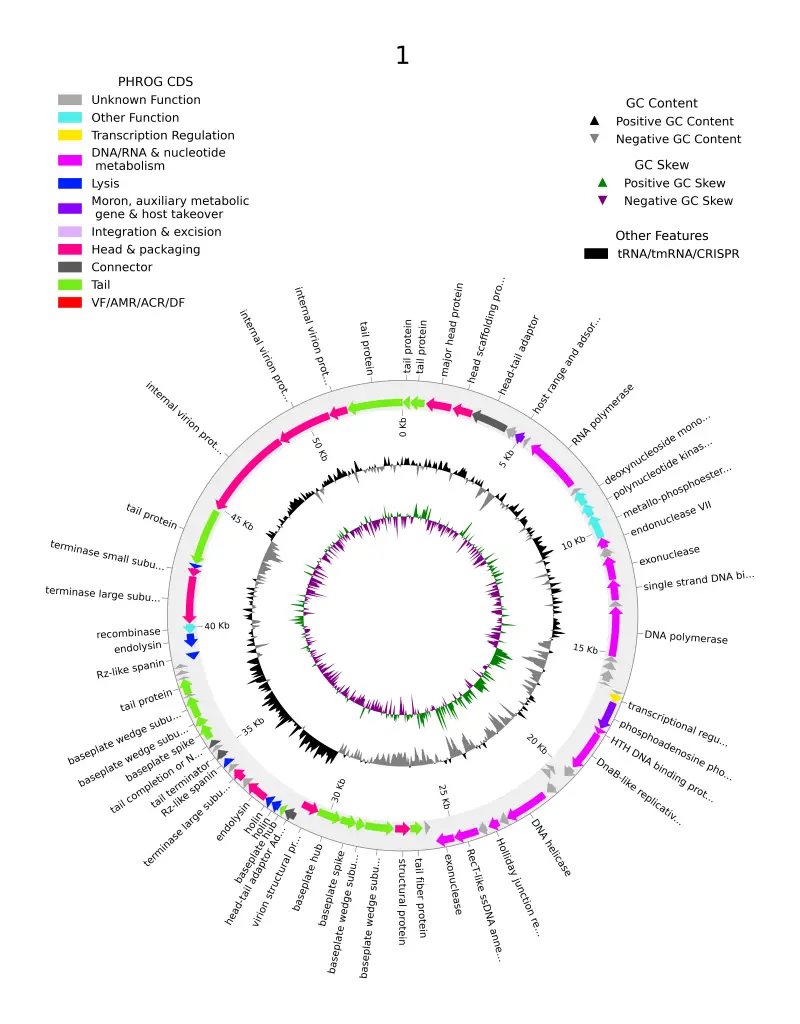
I know the majority of phage scientists have used Pharokka for phage annotation at one point. Despite being a very helpful tool, there were still large percentages of proteins with unknown functions. This challenge inspired the creation of another tool called PHOLD, developed by George Bouras who also created PHAROKKA. This cutting-edge tool is poised to transform the way scientists annotate phage genomes by predicting more features (and functions) using the protein morphology of proteins. But what makes Phold stand out in the crowded field of genomic tools?
Integrated with Pharokka
As both Pharokka and Phold are created by the same person, it seems that Phold was designed to complement Pharokka. Hence, Phold doesn’t work in isolation. Instead, it uses the GenBank file output from Pharokka as input, allowing researchers to refine and update annotations with greater precision. The slight downside is that you may need to have both tools installed, but Pharokka is also very simple to install and run.
The Backbone of Phold: Structural Homology
Traditional methods of genome annotation often struggle with the diverse and poorly characterized nature of phage proteins. However, Phold leverages the power of structural homology to enhance functional predictions. This approach relies on the principle that protein structure can provide more reliable clues about function than sequence alone. This tool employs the ProstT5 protein language model to swiftly convert protein amino acid sequences into the 3Di token format compatible with Foldseek. This process allows Foldseek to compare these tokens against a vast database of over one million phage protein structures.
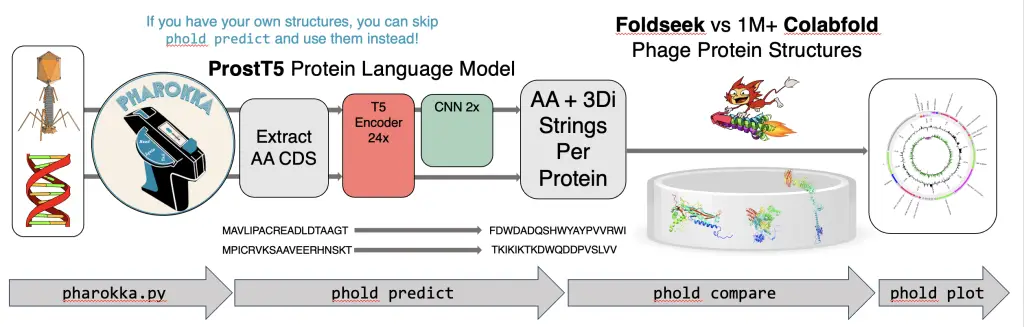
Cutting-Edge Technology Under the Hood
- ProstT5 Protein Language Model: Phold starts by predicting protein structures using the ProstT5 model. This advanced tool generates 3Di tokens from amino acid sequences, setting the stage for accurate structural comparisons.
- Foldseek for Structural Comparison: Once the protein structures are predicted, Phold uses Foldseek to compare these structures against its extensive database. Foldseek’s efficiency and accuracy in searching protein structures make it ideal for this task.
- Phold Database: The Phold database is a treasure trove having phage protein structures. Sourced from PHROG, anti-CRISPR, Defensefinder, VFDB, and CARD, this database ensures comprehensive comparisons and accurate annotations.
- Probabilistic Annotation: Phold goes a step further by incorporating probabilistic annotation. This approach evaluates the confidence in each functional prediction, offering a nuanced view of protein functions and highlighting areas of uncertainty.
User-Friendly and Efficient
Phold is designed with user convenience in mind. Installation is streamlined via Mamba, a package manager that simplifies dependency handling. Users with GPUs can benefit from accelerated performance, but CPU-only options are available for those without GPUs.
Just like its counterpart Pharokka, Phold involves straightforward single-line commands, making it accessible even for those new to bioinformatics. From installation to generating detailed visualizations like Circos plots, Phold’s workflow is intuitive and efficient.
Real-World Applications
Phold is especially beneficial for researchers investigating phages from diverse and underexplored environments, such as those working with metagenomic datasets. However, its efficiency extends to all phage researchers. By providing more accurate annotations, Phold significantly enhances our understanding of phage biology and their interactions with bacterial hosts. It marks a major advancement in phage genome annotation, setting a new standard with its innovative approach to structural homology, extensive database, and probabilistic annotation. For those exploring the complex world of phages, Phold is a powerful tool to try out.
For a deeper dive into Phold, including detailed tutorials and installation guides, visit the Phold GitHub page. Images used in this article are from the PHOLD Git-Hub page except the cover page which was from my work, when annotating my phage using PHOLD.

Leave a Reply
You must be logged in to post a comment.