Knowing exactly what your phages are and whether any of that kind already exists in the database is crucial for any phage scientist. However, achieving that can be as complex as navigating a maze or as simple as writing a single line of code, with the script doing all the work for you. You may not have heard of tax_myPHAGE yet, but that is precisely what it does: classifying your double-stranded DNA phages. This tool Developed by Andrew Millard, Thomas Sicheritz-Ponten and Remi Denise offers an efficient solution for assigning taxonomy to bacteriophages at the genus and species levels. It leverages current standards set by the International Committee on Taxonomy of Viruses (ICTV) and provides an automated process to determine the closest relatives of a given phage genome within the ICTV-classified genomes.
Key Features of TaxMyPhage
Purpose and Design:
- Specificity: TaxMyPhage is designed specifically for double-stranded DNA (dsDNA) phage genomes. It determines whether a phage belongs to an existing genus or species by comparing its genome against a database of ICTV-classified genomes. Additionally, the tool can identify if your genome represents a new species. However, if you use an input that isn’t a double-stranded DNA phage, such as RNA phage genomes or single-stranded DNA phage genomes, the script will still run but won’t produce accurate results.
- Automation: The tool automates the process of genome comparison, utilizing a VIRIDIC-like analysis to determine the taxonomic classification of the phage.
- Accuracy: By adhering to ICTV’s established cutoffs for genera and species, TaxMyPhage ensures that the classification is consistent with current standards.
Functionality:
- Genus and Species Identification: The tool identifies whether a phage genome fits within an existing genus or species and alerts the user if the phage represents a new genus.
- Multiple Input Support: Users can input multiple phage genomes simultaneously, making it easier to analyze large datasets.
- Database Updates: TaxMyPhage allows for manual updates of the databases to align with the latest ICTV taxonomy, ensuring ongoing accuracy and relevance.
Limitations:
- Scope: The tool is specifically designed for dsDNA phages and may provide inaccurate results for RNA phages or single-stranded DNA (ssDNA) phages. It does not classify metagenomic samples, or eukaryotic viruses, or assign phages to new families.
- Database Scope: TaxMyPhage compares genomes against currently classified ICTV phages rather than every phage genome available in GenBank, focusing on accurate classification rather than exhaustive comparison.
Installation and Quick Start
TaxMyPhage can be easily installed using Conda or pip. However, macOS users with M1/M2 chips will need to install specific packages before running the tool, as outlined in the installation guide. For more details, you can visit the GitHub page. The developers have also mentioned that a web version is coming soon, which will be particularly helpful for those who prefer to avoid the complexities of coding. That said, the current setup is straightforward: with Conda, it’s just three simple steps from installation to running the script.
To help you get started (for conda), here’s the code for installing and using the tool
Create and activate the environment:
mamba create -n taxmyphage -c conda-forge -c bioconda taxmyphage mamba activate taxmyphage
Install the necessary databases:
taxmyphage install
Run the analysis:
taxmyphage run -i test.fna -t 4
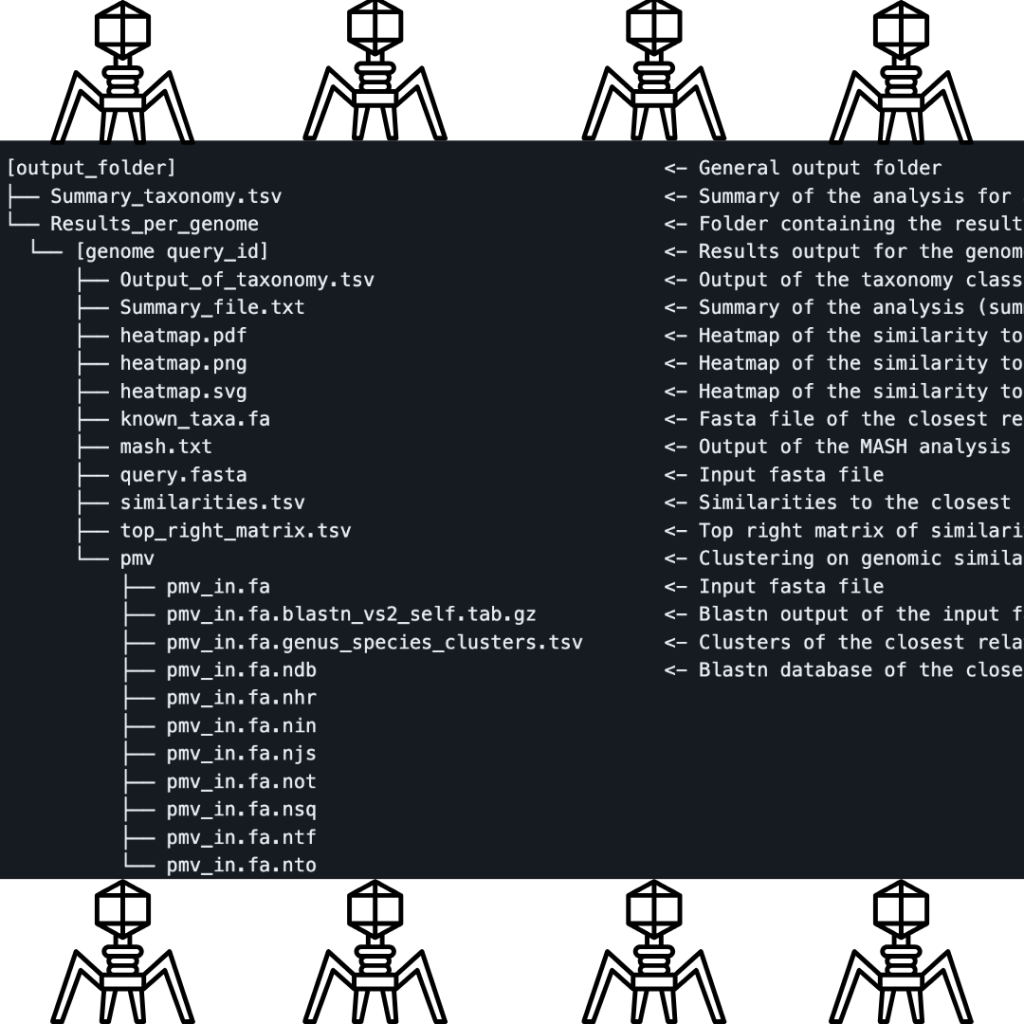
Output and Interpretation
The output of TaxMyPhage is easier to understand, as the tool is designed to produce a file with a structure similar to ICTV files, which simplifies the interpretation of the results. You can easily visit the ICTV website for more details regarding the group in which your phage is classified since the classification is based on ICTV standards. The output also includes a comprehensive summary of the classification process, heatmaps showing genomic similarity to the closest relatives, and detailed results for each genome analyzed, as your input sequence(s) may not necessarily match completely with what is in the databases. The key output files include:
- Summary_taxonomy.tsv: A summary of the analysis for all genomes.
- Heatmap files (.pdf, .png, .svg): Visual representation of the similarity between the query genome and its closest relatives.
- Output_of_taxonomy.tsv: A detailed classification of each phage, including the genus and species assignment.
Try out…..
If you haven’t yet tried TaxMyPhage, it’s worth considering, though it currently supports only double-stranded bacteriophages (which are the majority of sequenced phages). Perhaps in the future, the creators will expand its capabilities to include other types of bacteriophages. I’ve tried TaxMyPhage myself, and I liked it. Although I used it with metagenomic-mined phage genomes and, as expected, most of my phages had no hits, the tool provided a very detailed and clear classification for those that did. Utilizing ICTV standards and providing an automated workflow, significantly helps researchers avoid the confusion that often arises from phages being given random names by those who isolate them. Whether you’re analyzing individual genomes or multiple datasets, TaxMyPhage is an essential resource for anyone working in the field of bacteriophage research.
The cover image is sourced from the TaxMyPhage GitHub page, where you can find more information about the tool. To discover more phage-related bioinformatics tools, visit our bioinformatics category page or explore the related topics suggested below this article.

Leave a Reply
You must be logged in to post a comment.