Imagine you’re a scientist in the early 1900s, just discovering that tiny viruses can infect and destroy bacteria. You’re calling them “bacteriophages,” and it makes sense to name them based on their bacterial host. After all, if you find a virus that infects E. coli, why not call it an E. coli phage? Simple, right? But fast forward to today—our knowledge of these viruses has exploded, and things are far more complex. Does the simplicity of this host-based naming system still hold up, or is it time to embrace a more standardized, scientific approach?
We, along with the Quadram Institute communication team, recently held an interview with the Vice President of the International Committee on Taxonomy of Viruses (ICTV) Dr. Evelien M. Adriaenssens to discuss recent changes in phage taxonomy. I decided to prepare this blog post to explain what motivated me to conduct this interview with her. Let’s dive into the debate: simplicity vs. standardization in bacteriophage nomenclature. Which approach truly benefits science—and why does it matter?
Early Days: Simplicity Reigns
In the early 20th century, when scientists like Frederick Twort (1915) and Félix d’Hérelle (1917) discovered bacteriophages, they were working in a world where advanced molecular biology didn’t yet exist. Back then, the most practical way to identify these viruses was by their bacterial hosts. A phage that attacked Pseudomonas? Easy—call it a Pseudomonas phage. One that infected Salmonella? Let’s go with Salmonella phage.
This naming system was not only simple but also effective. If you knew what bacteria the phage infected, you had a good idea of what it was. It reflected the basic understanding of how these viruses worked—bacteriophages were specific to certain bacteria, so their names were tied to their hosts. It was all about keeping things straightforward.
But was it too simple?
The Turning Point: When Simplicity Became a Problem
As scientists like Max Delbrück, Alfred Hershey, and Salvador Luria began to study phages more deeply in the mid-20th century, they discovered something remarkable: phages weren’t just simple “bacteria eaters.” They were complex entities with genetic material, and they could be studied to understand broader biological processes—like how DNA functions as the carrier of genetic information. Hershey and Chase’s famous experiment in 1952, using T2 phage, forever changed our understanding of genetics.
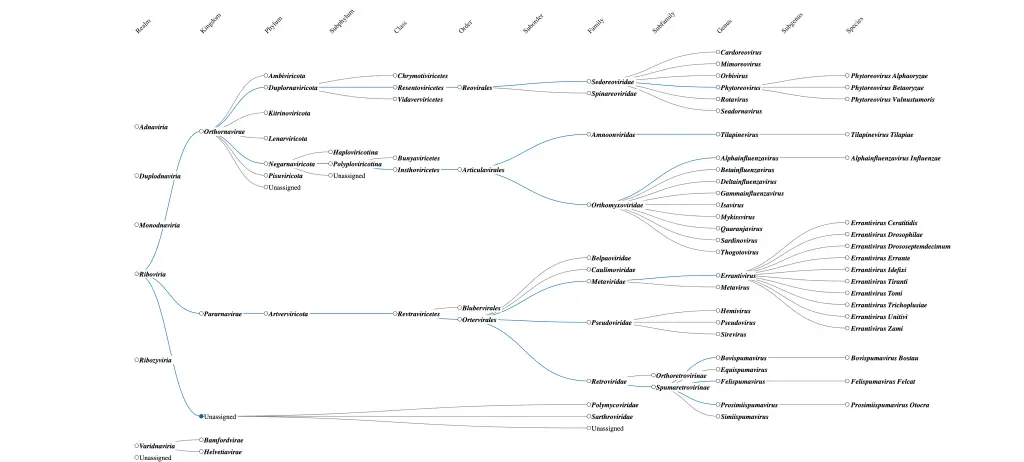
Suddenly, phages weren’t just tools for killing bacteria—they were windows into the molecular world. And naming them based solely on their bacterial hosts no longer captured this complexity. This is where the need for standardization came in. However, the standardisation that was adopted at that moment wasn’t standard enough as the structure/morphology became the basis of classifying these virus particles. Don’t get me wrong, this system worked for that time. For instance, families like Myoviridae, Siphoviridae, and Podoviridae were historically grouped based on their morphology, rather than solely on their host organisms. Interestingly, many phages that were once classified by morphology are still considered closely related in the modern, genomics-based classification.
The Rise of Standardization: A Modern Necessity
As phage research advanced, especially with the advent of molecular biology, it became clear that naming viruses after their hosts was no longer enough. With thousands of phages being discovered, many infecting the same bacteria but with different structures, replication mechanisms, or genetic makeups, a more systematic approach was required.
ICTV took on the challenge of creating a standardized, hierarchical classification system. Now, instead of simply naming a phage based on the bacteria it infects or their morphology, scientists classify phages based on:
- Genomic sequences: Is it a DNA or RNA virus? Genomic Sequence Similarity? What are the key genes? it’s phylogeny?
This level of detail allows scientists to better understand the relationships between different phages, their evolutionary history, and their broader biological significance.
But wait—does all this detail make things too complicated?
The Trade-off: Simplicity vs. Standardization
Let’s take a step back. Simplicity makes things accessible. It’s easy to remember that a “T4 phage” infects E. coli, or that a Salmonella phage goes after Salmonella. But simplicity can also be limiting. When phages are classified only by their hosts, we miss out on the fascinating complexity of their genetics, evolution, and diversity.
Standardization, on the other hand, offers precision. It ensures that we’re naming and grouping phages in a way that reflects their true biological nature. But with precision comes complexity. For someone new to virology, the ICTV’s classification system might feel overwhelming at first.
So, which is better?
The Best of Both Worlds?
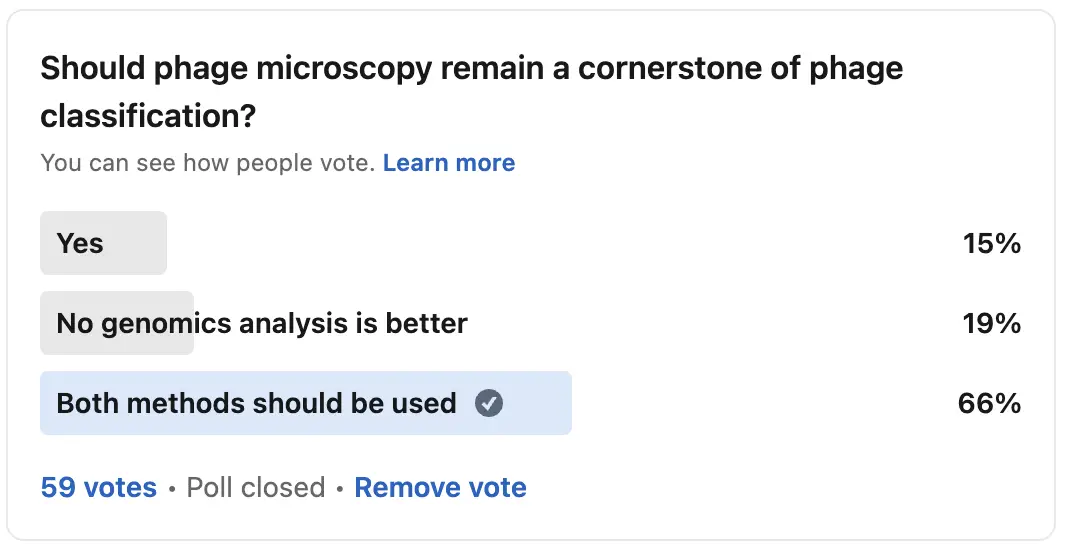
Perhaps the answer isn’t choosing between simplicity or standardization—it’s finding a balance. In the early stages of phage discovery, simplicity had its place. It was practical and effective in a world without advanced genomic tools. However as our understanding of these viruses deepened, the need for standardization became apparent.
Today, scientists benefit from both approaches. For example, we still use the host-based naming convention in everyday lab language (e.g., E. coli phages), while at the same time, the ICTV provides a rigorous classification system that captures the true complexity of bacteriophages. This combination allows researchers to communicate effectively while still delving into the finer details of phage biology.
Join the Debate: What’s Your Take?
What do you think—should we prioritize simplicity for the sake of accessibility, or is standardization the way forward in a world of increasing scientific complexity?
Perhaps, as with most things in science, there’s no one-size-fits-all answer. But understanding the evolution of bacteriophage nomenclature helps us appreciate how far we’ve come—and why striking the right balance is crucial for the future of virology. Watch our interview with the vice president of ICTV on Phage classification and the recent changes



Leave a Reply
You must be logged in to post a comment.