The study of bacteriophages, or simply “phages,” is one of the most fascinating areas of microbiology. These are viruses that infect bacteria, and they are everywhere — from the deepest oceans to the soil in your garden. Understanding phages at the genomic level is a bit like assembling a massive, intricate puzzle because they are so diverse, carrying unique genes that can be difficult to annotate. Additionally, phage genomes remain underexplored, meaning that some genes are not yet included in the databases used by sequence-based annotation tools. This is where Phynteny, a tool developed by Susie Grigson from Flinders University, comes in. It uses a language learning model to perform gene annotation.
Let’s break down what Phynteny is, why it matters, and how it helps scientists in the field of phage research.
What Are Phages and Why Study Them?
Phages are viruses that specifically infect bacteria. They look like tiny lunar landers, attaching themselves to a bacterium before injecting their genetic material into the host. Once inside, phages take over the bacterial cell’s machinery, forcing it to produce more phages until the cell bursts open, releasing new viruses.
The study of phages is incredibly important because they are everywhere, outnumbering all other forms of life on the planet. Scientists estimate that there are around 10³¹ phages on Earth — that’s more than every other living organism combined! They play a crucial role in maintaining the balance of ecosystems by controlling bacterial populations and are even being studied as alternatives to antibiotics in the fight against antibiotic-resistant bacteria.
The Challenge of Phage Genomes
Like many other viruses, phages have small genomes, with relatively few genes as they depend much on their hosts. But here’s the challenge: about 65% of these phage genes don’t match anything we know. Therefore, when trying to annotate them using sequence-based methods, these genes are labelled as ‘hypothetical proteins’ because their functions are unknown. In other words, they are genetic mysteries that scientists are eager to solve.
Traditional methods for annotating genomes — that is, figuring out what each gene does — are not always effective for phages because of the high number of unknown genes. Phynteny provides a new way to approach this problem by using a concept called synteny.
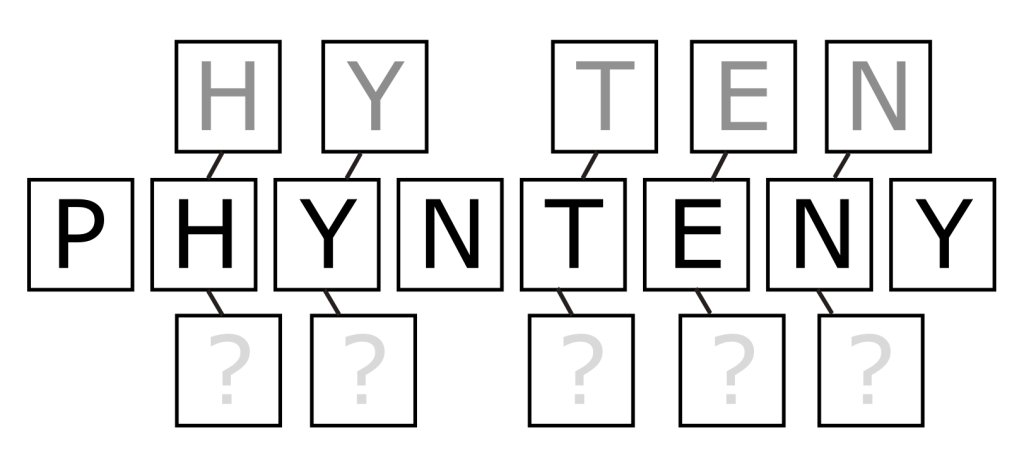
What Is Synteny and Why Is It Important?
Imagine you have two recipes from different cultures, but they both make the same dish. Even though the ingredients and instructions may differ, the order of steps might be similar because they’re trying to achieve the same result. Synteny in genetics works similarly. It refers to the preserved order of genes across different but related organisms.
Phages that are related — even distantly — tend to keep the order of their genes relatively stable over evolutionary time. This means that if scientists can identify a pattern in one phage, they might be able to predict what similar-looking genes are doing in another, even if those genes are technically unknown.
Phynteny uses this idea to make intelligent guesses about what hypothetical phage proteins might do, based on their position in the genome relative to other, known genes.
How Does Phynteny Work?
Phynteny is a computational tool that relies on a machine learning technique called Long Short-Term Memory (LSTM). LSTM is a type of artificial intelligence that is particularly good at recognizing patterns in sequences, like how sentences are structured in a language. In this case, it looks at gene sequences and their order.
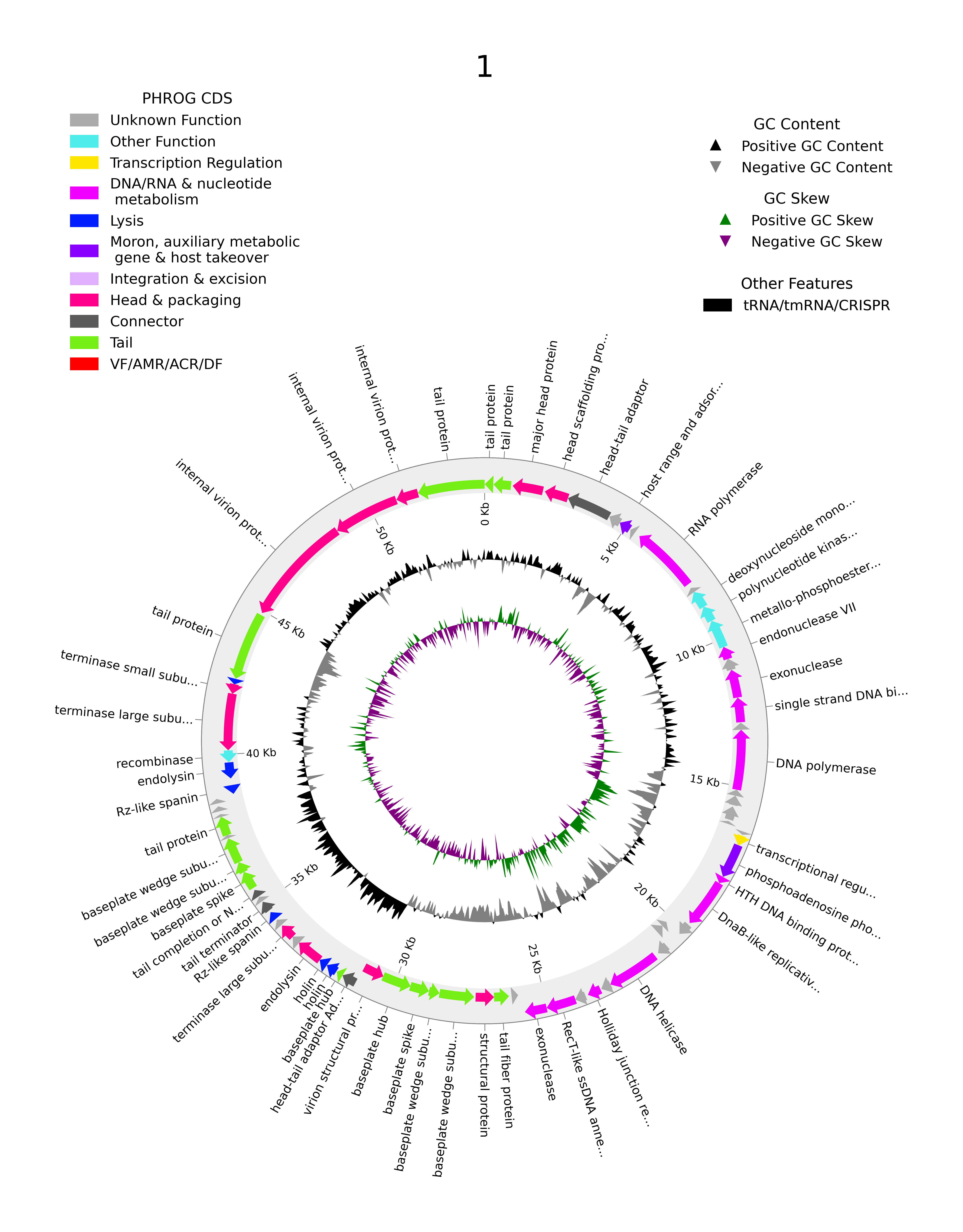
The tool uses a special dataset called the PHROG database (Phage Orthologous Groups), which is a collection of well-studied phage proteins grouped by their functions. Phynteny can match unknown genes in a new phage to similar groups in the database based on gene order and patterns, making it easier to predict their functions.
Making Phage Genomes Less Mysterious
Phynteny doesn’t just look at one gene at a time; it considers the context of the entire genome. This is important because genes often work together in networks or pathways. By understanding the “neighbourhood” that a gene lives in, Phynteny can make better predictions. If a hypothetical protein is consistently found next to genes that are involved in breaking down bacterial cell walls, for example, it’s a good bet that this unknown protein is also related to that process.
Why Does This Matter?
- Phage Therapy: One exciting area of phage research is the development of phage therapy — using phages to target antibiotic-resistant bacteria. Knowing what phage genes do can help scientists engineer or select phages that are more effective against specific bacterial infections.
- Environmental Impact: Phages are key players in ecosystems. Understanding their genomes can give us insight into how they regulate bacterial populations in various environments, from oceans to the human gut.
- Biotechnology: Phages are also valuable tools in biotechnology. They can be used to deliver genes into bacteria, which is crucial for developing new vaccines or creating bacteria that produce useful substances, like biodegradable plastics.
How Scientists Can Use Phynteny
For researchers interested in trying out Phynteny, the tool is accessible and adaptable. It can be installed through Bioconda or PyPI. Scientists can use pre-trained models or customize their data inputs, making it easy for both new and experienced users to apply the tool to their datasets.
Phynteny’s use of machine learning and gene synteny gives it an edge over traditional annotation methods, particularly when dealing with genomes that contain a high percentage of unknown genes. Its accuracy improves as more phage genomes are sequenced and studied, gradually building a more detailed map of the “dark matter” in phage genetics.
A Tool for the Future
The field of phage research is rapidly evolving, with more and more emphasis on understanding the unknown. Tools like Phynteny and Phold (Which use protein structure) are at the forefront, making sense of genetic information that was previously indecipherable. By turning hypothetical proteins into something worth exploring in the phage world. Phynteny is not just solving puzzles — it’s helping scientists study those annotated proteins while forming hypotheses about what these proteins are and what they do.
Phynteny is like a detective tool for phage researchers, using clues hidden in the genome’s structure to suggest the identities of mysterious genes. For anyone who is doing phage work, this is another step towards understanding how these minute viruses live.
For more information and to get started with Phynteny, visit the Phynteny GitHub repository.

Leave a Reply
You must be logged in to post a comment.