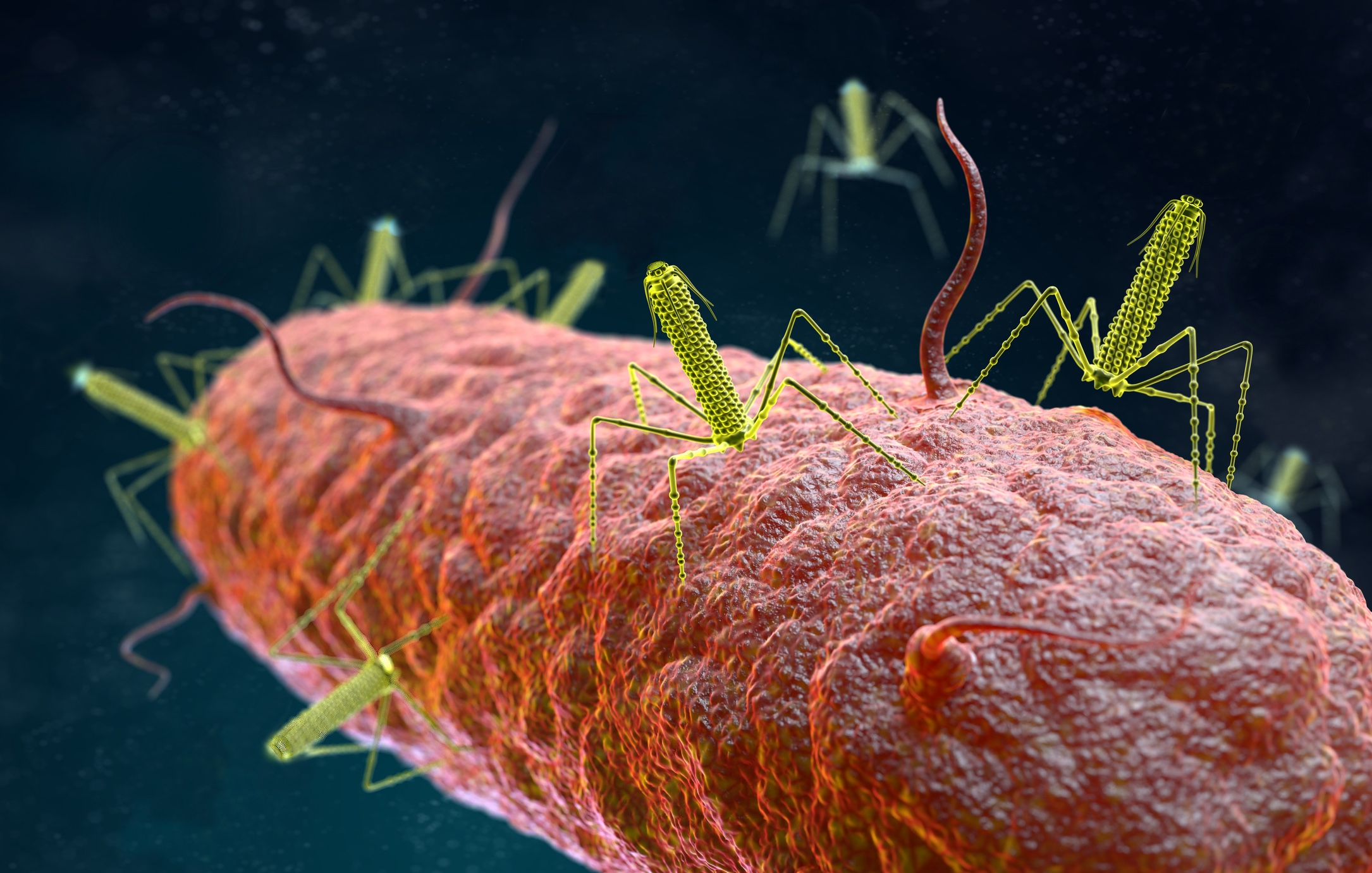
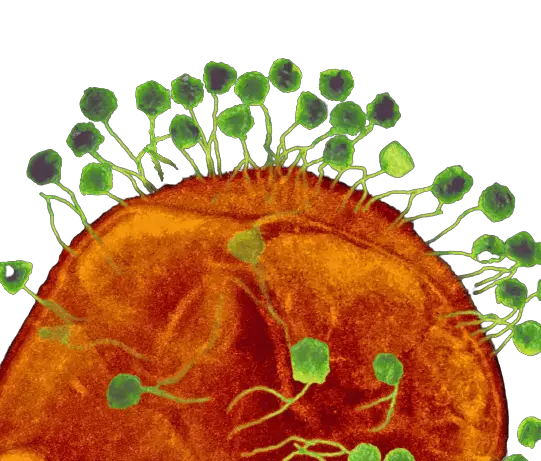
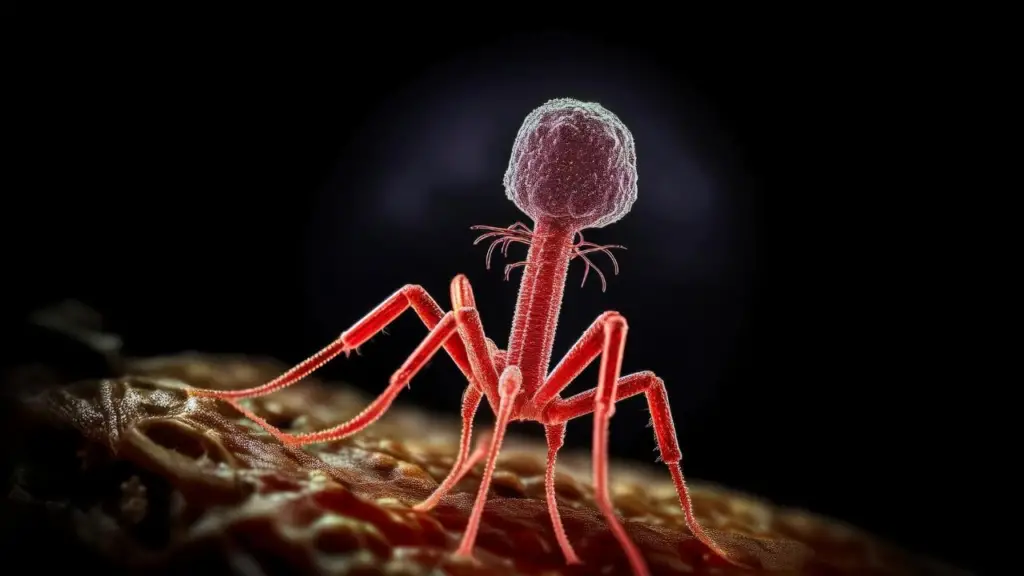
Factors governing phage replication and phage transmission in situ are unknown, especially within the confines of the gastrointestinal tract. We are unaware of the consequences of phage replication on the bacterial community and tissues of the animal host, and whether the phages are in the form of extracellular virions or intracellular phages.
Scientists from the University of California have created ‘Phollow’, which is a collection of tools and techniques to monitor phage outbreaks in situ by live imaging. Phollow is a ‘live image tracking’ with single virion resolution implemented to track phage replication and spread. This was achieved by marking the phages with distinct fluorescent proteins during assembly in newly infected cells.
The construction of Phollow phages
P2-like phages belonging to the caudoviricetes class of tailed phages, which infect bacterial genera of the phylum Proteobacteria, were chosen. Fluorescently marked ‘Phollow phages’ were constructed using the SpyTag: SpyCatcher tagging system, devised to mark the phage virions fluorescently with optically transparent zebrafish. However, they were marked by a different colour upon infection and replication within the new bacterial host. This combination labelling strategy is called ‘Phollow’.
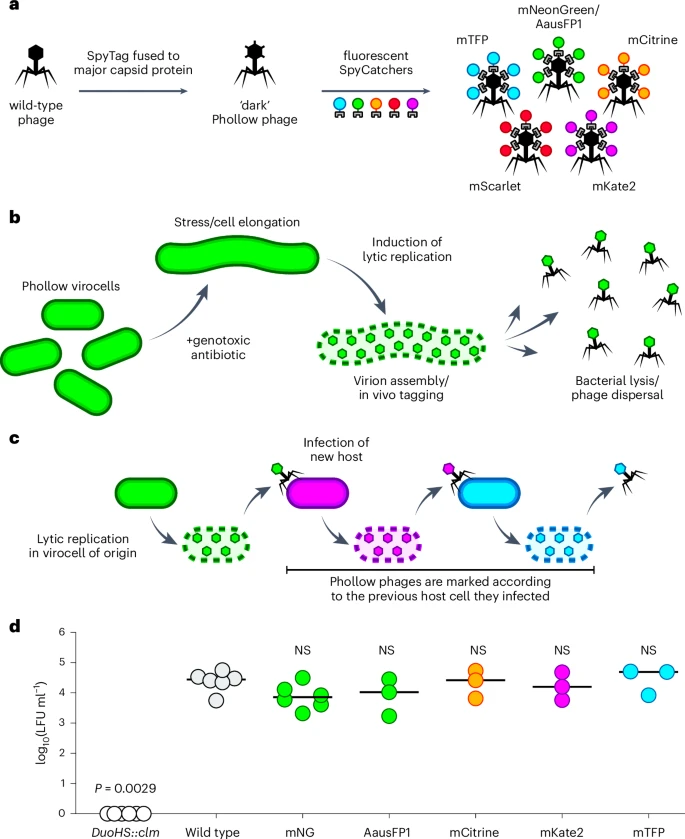
The in vivo observation
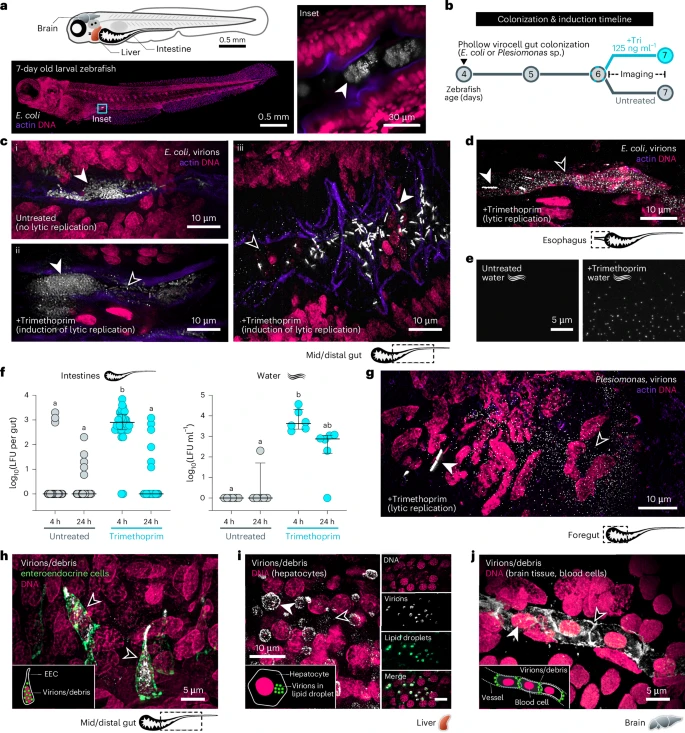
In vivo, colonized germ-free zebrafish were chosen with a two-member bacterial community composed of AausFP1 E. coli Phollow virocells, and mKate2 E. coli target cells. The viral replication was induced by using antibiotics, and phage activity was monitored using time-lapse imaging. ‘Phollow’ was used to compare the magnitude of virion dispersal induced by genotoxic antibiotics with different modes of action and minimum inhibitory concentrations (MIC). Flow virometry showed that mitomycin C, ciprofloxacin, and trimethoprim all induced peak virion output. Trimethoprim-induced outbreaks in zebrafish intestines produced clouds of phages. Plesiomonas-derived phages spread systemically to the liver and brain.
The in vitro study
In vitro, Phollow phages were used to quantitatively track phage transmission in a two-member community of mNG E. coli Phollow virocells and mKate2 E. coli target. Cells were found replicating in new host cells and undergoing onward interbacterial transmission. This experiment demonstrates how Phollow can reveal the underlying transmission dynamics that shape bacterial communities.
Phage engineering requires a more detailed understanding of the phage behaviour, especially interbacterial gene transmission, which has been technically more challenging. The solution for this can be ‘Phollow’, which blends new genetic tools and experimental models to investigate phage lytic replication and transmission in situ. Initial testing of Phollow shows its capability to enable multiscale investigations of phage transmission and transkingdom interactions that may open new avenues for phage-based microbiome therapies.
Read the article here https://www.nature.com/articles/s41564-025-01981-1
For more phage-related updates, click herehttps://www.thephage.xyz/category/news/