Simplified workflow to assemble and annotate phage genome
What is Phage Annotation?
Phage annotation involves identifying and labelling the functional elements within a phage genome. This includes coding sequences (CDS), tRNA genes, regulatory elements, and any other genomic features. The goal is to provide a complete and accurate map of the phage genome that can be used for further research, including phage therapy, genetic engineering, and evolutionary studies.





Importance of Phage Annotation
- Understanding Phage Biology: Annotated genomes reveal the roles of various genes in the phage lifecycle, host interaction, and replication mechanisms.
- Phage Therapy Applications: Accurate annotation helps in identifying genes that could be modified or utilized in phage therapy, an emerging alternative to antibiotics.
- Biotechnological Applications: Phages are tools in genetic engineering, and understanding their genome allows for precise manipulations.
Step-by-Step Process of Phage Annotation
- Starting with Raw SequencesThe annotation process begins with obtaining the raw sequence data of the phage genome. This can be done through high-throughput sequencing technologies, such as Illumina for short reads or Oxford Nanopore for long reads.
- Quality Control Using FastQCBefore any analysis, it’s crucial to ensure the quality of the raw sequencing data. FastQC is a widely used tool that provides a visual overview of the data quality, identifying issues like adapter contamination, low-quality reads, or unusual GC content. Quality control ensures that only high-quality data is used in downstream analyses, preventing errors and ensuring the accuracy of the final annotation.
- Genome AssemblyThe next step is assembling the reads into a contiguous sequence (contig). This is where the choice of tool depends on the type of data available:
- SHOVILL: Used for assembling short-read data, SHOVILL is a pipeline that streamlines the process of assembling bacterial genomes.Trycycler: Ideal for hybrid assembly of both short and long reads, Trycycler combines the strengths of different sequencing technologies to produce high-quality assemblies.
- Read Mapping with minimap2Once assembled, the genome needs to be validated and analyzed further. minimap2 is an efficient tool for mapping reads back to the assembled genome, allowing for the identification of any potential misassemblies and the validation of the assembly’s accuracy. This step ensures that the genome assembly is consistent with the original sequencing data.
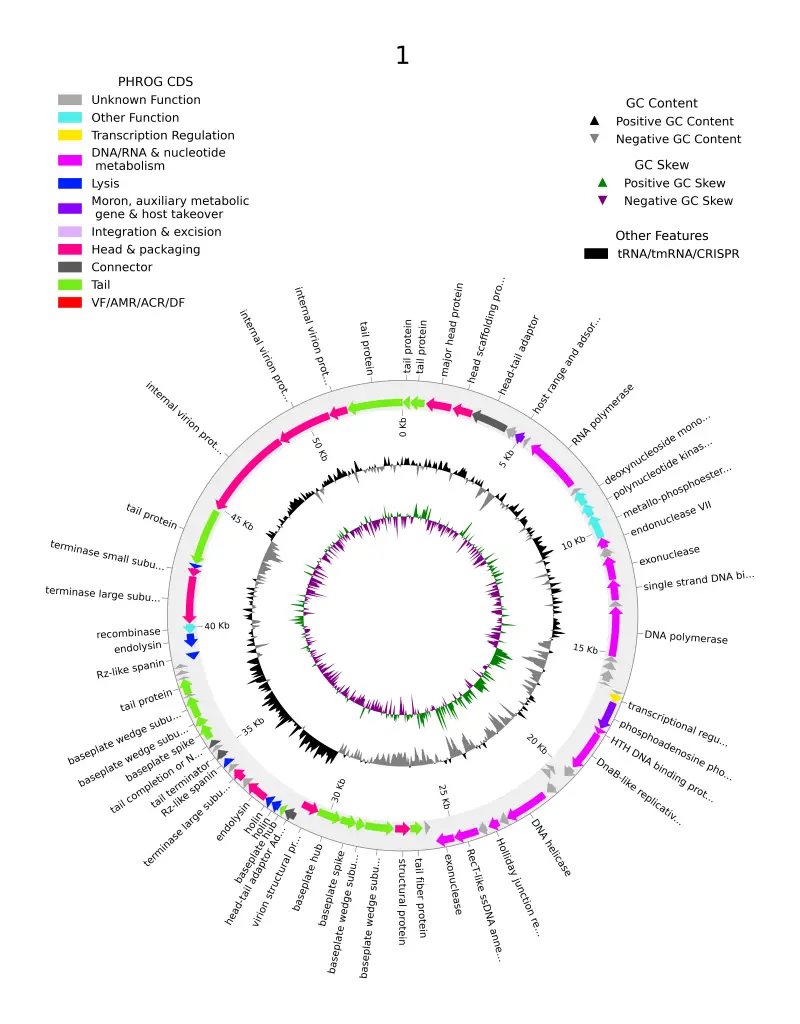
- Annotation Using PharokkaWith the genome assembled and validated, the next step is the actual annotation. Pharokka is a popular tool designed specifically for phage genome annotation. It identifies coding sequences (CDS), tRNA genes, and other genomic features, assigning functions based on homology to known proteins and sequences. Pharokka’s automated pipeline is tailored for phage genomes, making it a preferred choice for researchers focusing on bacteriophages.
- Further Annotation with PholdEven after using advanced annotation tools like Pharokka, some genes may remain unannotated. Phold steps in to fill this gap by using machine learning techniques to predict the function of these unannotated genes, offering a more complete annotation. Phold complements the initial annotation by providing additional insights and increasing the overall accuracy of the genome annotation.
Challenges in Phage Annotation
Phage genomes can be highly diverse and mosaic, which poses challenges in accurate annotation. Many phage genes have no known homologs, making functional predictions difficult. The reliance on databases that may lack phage-specific sequences also limits the accuracy of annotations. These challenges underscore the need for continued development of specialized tools and databases for phage annotation.
Future of Phage Annotation
The future of phage annotation lies in the integration of artificial intelligence and machine learning. These technologies have the potential to predict gene functions with greater accuracy and identify novel genes that traditional methods may overlook. Additionally, the growing interest in phage therapy is driving the development of more comprehensive phage databases, which will further improve annotation accuracy.
Conclusion
Phage annotation is a complex but essential process for understanding bacteriophages. With tools like FastQC, SHOVILL, Trycycler, minimap2, Pharokka, and Phold, researchers can achieve accurate and comprehensive genome annotations. As the field advances, these tools will continue to evolve, making phage annotation more accessible and accurate, ultimately contributing to the broader fields of virology, biotechnology, and medicine.
By following the steps outlined in this guide, you can ensure that your phage annotation projects are conducted with precision, leading to valuable insights into the fascinating world of bacteriophages.