The most often utilized bacteriophages in phage display are filamentous bacteriophages from E. coli (f1, fd, and M13), while T4, T7, and phage have also been used. On this page (Application of phage display), you may read more about the many uses of this technology.
 |
| Electron microscopy of filamentous bacteriophage. Credit: Petrenko, 2018 |
1. E.coli filamentous bacteriophages (f1, fd, M13)
A form of bacteriophages known as filamentous bacteriophages are distinguished by their rod- or filament-like shape. Typically, filamentous phages attack Gram-negative bacteria and have a single-stranded DNA genome. Important phages that are useful in phage display include the M13 phage and members of the Ff, M13, fd, and f1 families.
The M13 bacteriophage is cylindrical in shape, measuring 880 nm in length and 6 nm in diameter. It contains a single-strand genome that encodes two categories of capsid proteins, major coat proteins (pVIII), and minor coat proteins, totaling five distinct capsid proteins (pVII, pIX, pVI and pIII).
Phage display frequently employs filamentous bacteriophages from E. coli. The majority of antibodies and peptides are visible at the pIII and pVIII phage proteins, which were used to build the pIII and pVIII display system. Additionally, the hybrid phage system allows for the visualization of large proteins that contain all five M13 coat proteins as N-terminal fusions with pIII, pVIII, pVII, and pIX as well as C-terminal fusions with pVI, pIII, and pVIII.
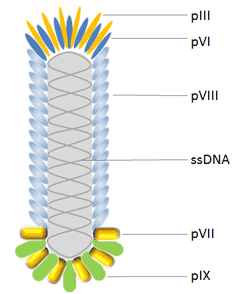
The protein known as pIII controls how infectious a virion is. It has 406 amino acid residues and appears in 3 to 5 copies at the phage tip. When utilizing a phagemid in conjunction with a helper phage, pIII allows for monovalent display, which is a benefit over pVIII. Additionally, pIII is more tolerant of it than pVIII and permits the insertion of longer protein sequences (>100 amino acids).
The primary coat protein of Ff phages is pVIII, which is produced in 2700 copies and is expressed by gene 8. As a result, it is employed to increase the detection signal when an antibody shown on a phage binds to an antigen. The N-terminus of Pviii, which is typically 6-8 amino acids long, is fused with peptides in most cases. Because of this, using this protein makes it difficult to find high affinity binding partners. Additionally, changes are made to pVIII to improve the effectiveness of the display; this has advanced significantly in recent years.
Due to its high throughput capability, pVI has been widely employed for the display of cDNA libraries and is a desirable alternative to the yeast-2-hybrid approach for the identification of interacting proteins and peptides. pVI has been chosen over pVIII and pIII for the production of cDNA libraries because the protein of interest can be added to pVI’s C-terminus without significantly altering its function in phage assembly.
In order to further enhance phage display as the top combinatorial engineering platform, pVII and pIX, which are positioned near the phage tip opposite that of pIII, may both complement existing phage display systems and be employed as alternate scaffolds for display and selection.
2. T4 Phage
Bacteriophage T4 (Enterobacteria phage T4) attacks the E. coli bacterium. It belongs to the T-even phage family, which also includes the enterobacteriophages T2 and T6. The T4 phage is a large phage, measuring around 200 nm in length and 90 nm in width. Its 169 kbps long double-stranded DNA genome contains 289 proteins. Three crucial proteins are used to construct the T4 phage: gp23, which creates the hexagonal lattice of the capsid, gp24, which forms pentamers at eleven of the twelve vertices, and gp20, which creates the special dodecameric portal vertex through which DNA enters during packaging and exits during infection.
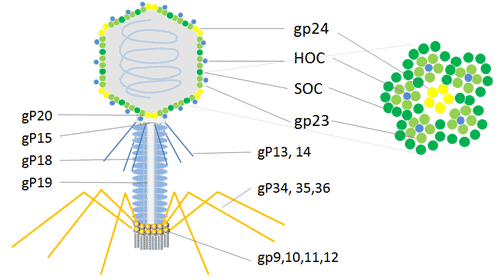
The T4 capsid is embellished with two non-essential outer capsid proteins, HOC (highly antigenic outer capsid protein) and SOC, in addition to the necessary capsid proteins, gp23, gp24, and gp20 (small outer capsid protein). Both HOC and SOC are dispensed with; they bind to the capsid following capsid elaboration. The production, viability, or infectiousness of phages are unaffected by null (amber or deletion) mutations in one of the genes or both of them.
For the phage display of numerous copies of peptides and proteins, the T4 capsid proteins HOC and SOC can be utilized. For the expression of cDNA and the display of peptides or proteins at high copy numbers on the phage capsid surface, the phage T4 HOC, SOC bipartite display system is appealing. It can be used to express cDNA, exhibits bigger proteins in high copy numbers, and inserts into the stop codon on the C-terminal of the 810-copy SOC protein or the 155-copy HOC protein[13]. In order to conduct research and produce immunological goods, the phage T4 dual-site display emerges as an effective technique with an improved immune response in animals.
3. T7 Phage
The Podoviridae family’s icosahedral T7 phage virus contains a linear double-stranded DNA genome. Bacteriophage T7 has a head and tail structure, just like T4. T7’s dsDNA genome is conserved in the icosahedral head, which has 415 copies of the capsid gp10 organized as 60 hexamers on the surface and 11 pentamers at the vertices. The main coat protein gp10 has two isoforms, gp10A and gp10B, which were created due to a spontaneous translational frame shift at amino acid 341.
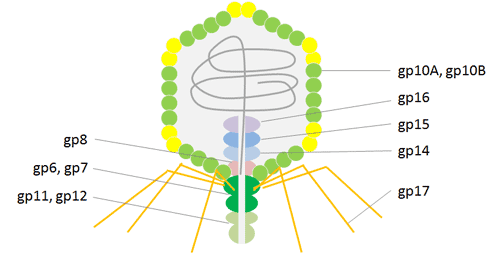

A frame shift at the gene’s terminus, which lengthens the capsid protein by 52 residues, causes the minor protein gp10B to be produced. The 52 additional residues on protein gp10B’s C-terminus are used to display fusion proteins. in order to prevent issues brought on by steric hindrance. It is possible to effectively carry out high throughput affinity elutriation using T7 phage particles due to their great stability under a variety of harsh conditions, such as high temperature and low pH. Recent uses of the T7 phage display technology, particularly in the areas of antigen identification, vaccine development, protein interaction, and cancer diagnosis and treatment, have been critical in revealing processes of molecular interaction.
4. Lambda phage
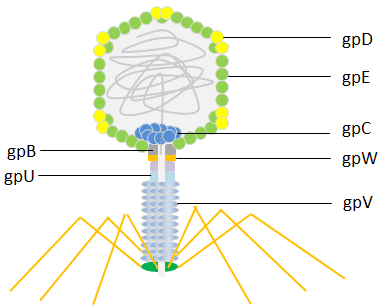
E. coli is infected by the bacterial virus known as lambda phage (also known as Enterobacteria phage or coliphage). Esther Lederberg made the discovery of it in 1950. The head of the lambda phage is icosahedral. The capsid protein gpD (402-420 copies) stabilizes the principal coat protein gpE (415 copies), which is used to build the shell’s primary structural components. The main tail protein gpV’s disks form a flexible helical tail that connects to the head.
Phage display has employed both the head protein gpD and the tail protein gpV. The lambda foo vector was initially built with a modest display level that made it suited for recording high-affinity contacts for the C-terminal display on gpV. Later, techniques for the display of peptides N- or C-terminally to the main coat protein gpD were created. The lambda phage is a potent tool for screening libraries of peptides, proteins, and cDNA because it has been made to efficiently display multiple copies of peptides or even large protein domains.
 read more about procedure of phage display here
read more about procedure of phage display here

Comments