As someone who has conducted bacteriophage research, I understand how common it is to encounter host DNA contamination when isolating a phage genome. This issue often only becomes apparent after completing sequence analysis, as tools like Nanodrop measure total DNA without indicating contamination. Given the high cost of sequencing in some regions, discovering contamination can be disheartening. While some kits provide standardized, ready-to-use methods for a fee—often significant in certain contexts—I felt it was important to explore low-cost alternatives. Researchers in similar situations might be struggling to access the same information.
Bacteriophages have genetic material that can be RNA or DNA, which may be circular or linear, single- or double-stranded. Isolating phage DNA is similar to isolating DNA from other microbes, though contamination from bacterial genetic material is a risk. Because phage isolation requires a large quantity of phages (measured in PFU), it may be necessary to multiply stocks. The enrichment process is used to increase the number of bacteriophages. In this approach, a culture of the target bacteria is grown overnight (depending on the host) and then mixed with the phages. This step carries the risk of contamination, as any lytic phages present in the sample can infect and multiply in the target bacterial culture.
Filtration to remove bacterial cell debris
After multiplication, the bacteria and other debris are removed through filtration. Filters with pore sizes of 0.22 and 0.45 microns are commonly used in this step. These filters can be either syringe adapter filters or filter paper combined with a chamber and a suction pump. The latter method is typically used for larger sample volumes, as syringe adapter filters rely on manual pressure from your palm to push the liquid through.
Filtration effectively prevents large bacterial cells from passing through the filter, thus separating phages (in the filtrate) from bacteria (in the residue). However, since DNA molecules are small enough to pass through the pores, this step does not guarantee that bacterial genetic material is entirely removed.
Have you ever wondered why environmental DNase doesn’t degrade bacterial DNA? I had the same question, only to find that existing research already demonstrates the persistence of DNA in harsh environments. One possible explanation is that bacteria can “absorb” free-floating foreign genes—something we’ve all heard of. This suggests that the enzyme might not target a specific molecule. Therefore, my conclusion is that DNase won’t always degrade genetic material in the environment, particularly on surfaces like tubes, where the enzyme might not even be present. To ensure proper degradation, it’s best to introduce your own commercially available DNase.
The modified phenol-chloroform-isoamyl alcohol method
The modified phenol-chloroform-isoamyl alcohol method, as used by Nale et al., (2015) is the technique I thought would be useful here. Well! It may not be as cheap as the boiling method, but it produces the best results and may be significantly less expensive than other commercial kits. In molecular biology, chloroform extraction is a liquid-liquid extraction technique used to purify nucleic acids while removing proteins and lipids. Both phenol and chloroform are non-polar solvents. Water, on the other hand, is a very polar solvent. These liquid properties are the fundamental principles of extracting DNA using this method (we won’t go deep into that). But let’s dive into the procedure itself.
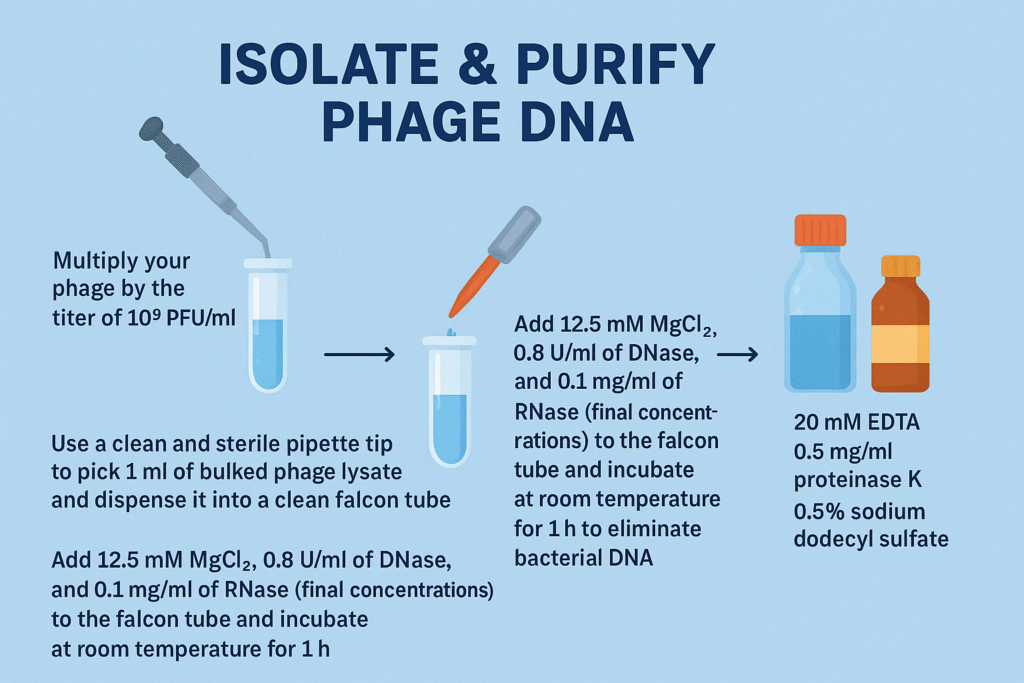
The procedure of extracting phage genomic DNA using the modified phenol-chloroform-isoamyl alcohol method
- Multiply your phage by the titer of 109 PFU/ml
- Use a clean and sterile pipette tip to pick 1 ml of bulked phage lysate and dispense it into a clean falcon tube.
- Add 12.5 mM MgCl2, 0.8 U/ml of DNase, and 0.1 mg/ml of RNase (final concentrations) to the Falcon tube and incubate at room temperature for 1 h to eliminate bacterial DNA.
- Add 20 mM of EDTA, 0.5 mg/ml of proteinase K, and 0.5% of sodium dodecyl sulfate before incubating at 55°C for an additional hour to digest the phage capsid. EDTA is a chelating agent; it removes the Mg ion, rendering the DNase inactive due to a lack of this divalent cation. Without this, your phage DNA will undoubtedly be digested.
- Mix an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and centrifuge at 15,000 × g for 5 min to extract the DNA.
- To precipitate the DNA, treat the aqueous layer obtained with 0.3 M sodium acetate and 2 volumes of ice-cold ethanol.
- After incubating for 10 min on ice, elute the DNA by centrifugation at 21,000 × g for 20 min.
- Wash the resultant pellet with 0.5 ml of 70% ethanol.
- Dissolve the pellet in 5 mM Tris-HCl. Boom! There you have your phage DNA
- Quantify and analyze your DNA (Nanodrop thing……………..)
For many other protocols click Here
References
- Nale J. Y., Spencer J., Hargreaves K. R., Buckley A. M., Trzepinski P., Douce G. R., et al. (2015). Bacteriophage combinations significantly reduce clostridium difficile growth in vitro and proliferation in vivo. Antimicrob. Agents Chemother. 60 968–981. 10.1128/AAC.01774-15.
- Grami, E., Badawy, S., Kiljunen, S., Saidi, N., & Skurnik, M. (2023). Characterization and genome analysis of Escherichia phage fBC-Eco01, isolated from wastewater in Tunisia. Archives of Virology, 168(2), 1-10.
- Ji, Y., Xi, H., Zhao, Z., Jiang, Q., Chen, C., Wang, X., … & Gu, J. (2023). Metagenomics analysis reveals potential pathways and drivers of piglet gut phage-mediated transfer of ARGs. Science of The Total Environment, 859, 160304.
- Chernyshov, S. V., Tsvetkova, D. V., & Mikoulinskaia, G. V. (2023). A rapid and efficient technique for the isolation of Bacillus genomic DNA using a cocktail of peptidoglycan hydrolases of different type. World Journal of Microbiology and Biotechnology, 39(1), 1-10.

Nice piece, more like a story and easier to understand
Is there an incubation step after adding the EDTA to allow chelation before adding the proteinase K?
[…] assembly is a method of genome assembly that involves piecing together overlapping fragments of DNA without the use of a reference genome. This method can be particularly useful for assembling phage […]
[…] size could allow it to pass through the filter pores. Therefore, it may be necessary to incorporate a nuclease step to digest free-floating nucleic acids before proceeding with the extraction of phage…. This precaution ensures a more precise and uncontaminated isolation of the desired genetic […]